-
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a potent and prevalent nitrosamine procarcinogen found in cigarette smoke. The aim of this work is to study alterations in peroxiredoxin (Prx) expression induced by NNK during carcinogenesis. Characterization of Prx genes from hamster was performed using bioinformatics. V79 cells were induced with different concentrations of NNK (0.1-0.4 mg/mL), and the expression levels of six Prx genes (Prx1-Prx6) were measured by qRT-PCR 24 h following NNK treatment. Prx gene expression was induced by NNK stress, and the highest transcription levels were induced by over 20.42-fold relative to that of the control. NNK induced alterations in Prx expression over the course of lung cancer, which means Prxs may play important roles in ROS detoxification under NNK stress and their functions are complementary.
Tobacco use results in over 400, 000 deaths annually in the United States, 157, 000 of which are due to lung cancer[1]. Besides exerting a direct oxidant effect, cigarette smoke also activates inflammatory cells to produce large amounts of reactive oxygen species (ROS) in the lungs. 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a potent and prevalent nitrosamine procarcinogen found in cigarette smoke, has been used extensively to study tobacco carcinogen-induced lung tumors in rodents and revealed to stimulate the survival and proliferation of lung cancer cells. Many studies have indicated that the 5-year survival rate of patients with lung cancer is closely related to the time of diagnosis. Unfortunately, most patients with lung cancer are diagnosed with advanced stages of the disease because of the lack of effective biomarkers or indicators for early diagnosis of lung cancer. Therefore, exploring effective biomarkers for the early diagnosis of lung cancer is important[2].
Mammalian cells feature well-defined endogenous antioxidant enzymes such as superoxide dismutases, catalase, and peroxiredoxins (Prx); among these enzymes, Prx is the most abundant. Prx scavenges ROS and influences diverse cellular processes, including growth, differentiation, apoptosis, and carcinogenesis[3]. These enzymes are classified into three major subclasses: typical 2-cysteine Prxs (Prx1-Prx4), atypical 2-cysteine Prxs (Prx5), and 1-cysteine Prxs (Prx6). Prx isoforms show distinct intracellular distributions and are localized in the cytosol (e.g., Prx1, Prx2, and Prx6), mitochondria (e.g., Prx3 or Prx5), endoplasmic reticulum (e.g., Prx4), and peroxisome (e.g., Prx5). Because of their structural characteristics, Prxs present distinct functions. For instance, 2-Cys Prxs, including Prx1-Prx4, have been indicated in multiple oncogenic signaling pathways and may thus contribute to various processes of cancer development. Knockout of the Prx1 gene causes spontaneous tumor formation in aging mice, which means Prx1 could function as a tumor suppressor. Prx2 plays an important role in reducing the deleterious effects of ROS, protects cells from oxidative damage, and increases the survival rate of cells. Prx3 acts as a mitochondrial scavenger of ROS, protects mitochondria against oxidative damage, and influences diverse cellular processes, including growth, differentiation, apoptosis, and carcinogenesis. The Prx family is highly expressed in human lung carcinoma, and various Prx members exert different effects on tumor progression[4]. To date, the alteration of Prx expression induced by environmental carcinogens such as NNK remains largely unknown.
Previous studies have indicated that antioxidant activity can be impaired by production of excessive oxidative stress in lung cancer, and oxidative stress is often associated with the altered expression of genes involved in ROS metabolism in cancer cells[5]. Thus, detailed analyses of the alterations in expression associated with redox reactions during inflammation are important in understanding the process of carcinogenesis and may present significant therapeutic implications. Considering that lung cancer is largely caused by tobacco smoke carcinogens such as NNK and that the majority of all lung cancer tumor tissue extracts express high levels of Prxs, our study investigates the association between these two molecules. In this study, we used qRT-PCR and Western blot to detect the expression of NNK-induced Prxs in V79 cells. Our results showed that NNK induces expression alterations in Prx genes over the course of lung cancer, which means Prxs may play important roles in ROS detoxification under NNK stress and be compensatory functionally.
NNK was purchased from Toronto Research Chemicals (North York, ON, Canada) and dissolved in DMSO to yield a saturated solution. The Chinese hamster lung fibroblast cell line V79 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured as previously described for 3 d to obtain growth-arrested cultures. To determine the effect of NNK on cell phenotype changes, growth-arrested cells were exposed to 0.1, 0.2, 0.3, 0.4, or 0.5 mg/mL NNK for 24 h. Cell viability was then examined by the trypan blue exclusion test.
Multiple alignment was conducted by CLUSTALX v1.81 using putative Prx genes identified from hamster. To reconstruct the phylogenetic tree of Prx genes, the protein sequence of the candidate gene was used as the query sequence to search for homologous sequences of other species in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/). The phylogenetic tree was reconstructed by MEGA4.0. Prediction of subcellular protein localization was performed for all identified Prx genes using PSORT (http://psort.ims.u-tokyo.ac.jp/form.html). Theoretical pI (isoelectric point) values and molecular weights were also computed for the genes via the ExPASy server (http://web.expasy.org/compute_pi/).
Total RNA was extracted from control and treated cells using Trizol reagent (Invitrogen) and treated with DNase I (Takara, Kyoto, Japan) to remove genomic DNA contamination. The primer pairs corresponding to Prx1-Prx6 and internal reference gene GAPDH are listed in Supplementary Table S1 (available in BES online, www.besjournal. com). Fluorescent qRT-PCR was carried out using a Real-time PCR Detection System (StepOnePlus, Applied Biosystems, Foster City, CA, USA) with the SYBR Premix EX Taq kit (Takara). At least three individual replicates were prepared for each sample, and each sample was analyzed three times. Analysis of Prx protein expression in V79 cells was conducted by Western blot.
The data were expressed as mean ± SD, and SPSS 12.0 (Chicago, IL, USA) was used for statistical analysis. The least significant difference (LSD) test was used to examine differences among means of the treatments and control (Supplementary Table S2, www.besjournal.com).
Six isoforms of Prx (Prx1-Prx6) have been identified in Cricetulus. The encoded ORFs deduced polypeptides of 198-274 amino acids with predicted molecular masses of 21.81-31.15 kD and pI values of 5.35-9.35 (Supplementary Table S1). Based on the number and position of conserved Cys residues, the Prxs were classified into three different types: Prx1-Prx4 were considered typical 2-Cys Prx proteins, Prx5 was considered to be an atypical 2-Cys Prx protein, and Prx6 was considered to be a 1-Cys Prx protein (Supplementary Figure S1, available in BES online, www.besjournal.com). Moreover, two conserved motifs, GGLG and YF, were identified in Prx1-Prx4; these motifs are critical signal transmitters that function through sensitive recognition of H2O2 changes. Prx5 and Prx6 did not show these conserved motifs. Prediction of subcellular localization indicated that Prx4 had a predicted signal peptide of 38 bp (Supplementary Figure S2, available in BES online, www.besjournal. com); thus, it is a secreted protein. While Prx1, Prx2, and Prx6 were located exclusively in the cytosol, Prx3 was located in the cytosol, mitochondria, and peroxisomes (Supplementary Table S1, available in BES online, www.besjournal.com). All these results reveal that the functions of Prx genes are fairly divergent. To investigate the evolutionary relationship between Cricetulus and mammalian genes, a phylogenetic tree of Prxs from Cricetulus, humans, rats, and mice was generated using a neighbor-joining method. Here, results suggested that most of the genes shared orthologous relationships between Cricetulus and mammals.
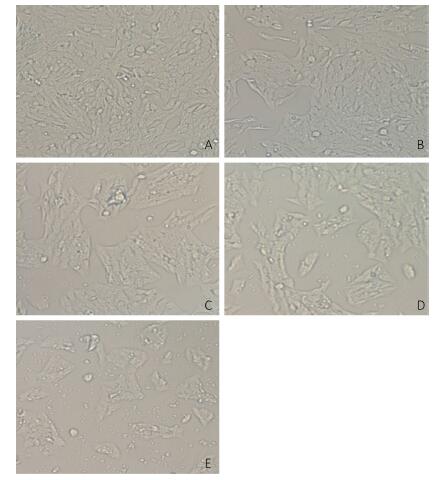
V79 cells were treated with different concentrations of NNK (0.1, 0.2, 0.3, or 0.4 mg/mL) for 24 h. All of the cells in the treatment and groups were then observed under an inverted microscope. Cells treated with 0.1 or 0.2 mg/mL NNK showed no significant changes in morphology compared with that of the control group. However, after treatment with 0.3 mg/mL NNK, the cell gap and numbers of dead cells increased while the number of living cells and growth density decreased. Treatment with 0.4 mg/mL NNK caused cell connections to nearly completely disappear, most cells to fall off, and more cell debris (Figure 1).
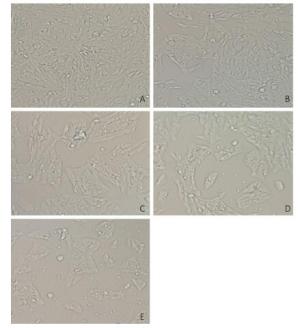
Figure 1. Effect of NNK on the morphology pf V79 cells 24 h after exposure (200 × ): (A) 0 mg/mL NNK, (B) 0.1 mg/mL NNK, (C) 0.2 mg/mL NNK, (D) 0.3 mg/mL NNK, and (E) 0.4 mg/mL NNK.
The expression profiles of the Prx genes were detected using qRT-PCR after V79 cell culture under normal conditions. The highest expression level of Prx4 mRNA was detected under normal conditions. Compared with that of Prx4, the expression levels of Prx1, Prx6, Prx5, and Prx3 mRNA gradually decreased; the expression of Prx2 was the lowest among these genes under normal conditions (Figure 2). These results suggest that the roles of Prx4, Prx1, Prx6, Prx5, Prx3, and Prx2 may gradually decrease during normal physiological and metabolic processes.
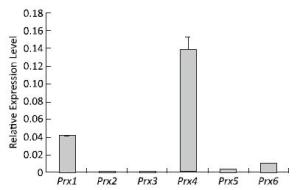
Figure 2. Expression patterns of Prx genes in V79 cells under normal conditions. Bars represent SD, and data points reflect the means of three independent experiments.
To study the relationships between Prx genes and their responses of NNK stress in V79 cells, the expression patterns of the genes in response to different concentrations of NNK were investigated by qRT-PCR and Western blot. After treatment with NNK, the expressions of all of the Prx genes were induced, and the highest transcription levels were over 20.42-fold greater than that of the control. The genes displayed different expression patterns and showed similar responses to different concentrations of NNK. These results suggest that Prxs may play important roles in ROS detoxification under NNK stress and be compensatory functionally. In a previous study, the Prx family was found to be highly expressed in human lung carcinoma. In particular, Prx2 is highly expressed in lung carcinomas. Overexpression of Prx2 by gene transfection induces cell proliferation and protects cells from apoptosis in vitro; the gene is also identified as aberrantly increased in human lung cancer and its levels are positively correlated with high-grade lung carcinomas[6]. However, the molecular basis of the contributions of Prx2 to lung cancer development has not been investigated. In our study, the expression of Prx2 was determined to aberrantly increase (20.42-fold) compared with that of the control after induction with 0.3 mg/mL NNK; transcription levels of Prx2 also showed an initial increasing trend, followed by a decrease, and then an increase. The expression level of Prx2 determined by Western blot analysis was similar to its expression at the transcriptional level (Figure 3); these findings highlight the importance of this gene in cancer and cancer progression in vitro.
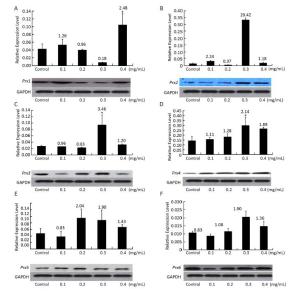
Figure 3. Expression of (A) Prx1, (B) Prx2, (C) Prx3, (D) Prx4, (E) Prx5, and (F) Prx6 in V79 cells after NNK induction. Bars represent SD, and data points reflect the means of three independent experiments. Values above bars indicate fold-increases in inducible transcription level relative to that of the control.
Prx1 is frequently identified as a major cellular antioxidant that is preferentially expressed in cancer tissues[7]. The gene mediates pro-oxidant-induced lung cancer cell growth and is required for human lung cancer cells to grow as tumor xenografts and establish cancer metastasis in mice[8]. We found that the expression of Prx1 increased after treatment with 0.4 mg/mL NNK by about 2.48-fold relative to the control. Transcription levels of Prx1 initially showed an increasing trend, decreased, and then increased once more.
Mitochondrial ROS can easily diffuse to the cytosol through both the inner and outer mitochondrial membranes[9], thereby inducing the response of cytosolic Prxs to oxidative stress in cancer. As an active responder to oxidative stress, Prx3 (alone or together with Prx5, another mitochondrial Prx gene) has been shown to be significantly upregulated in most common malignancies, including lung cancer[10]. Our results are consistent with this previous study and showed upregulation of Prx3 and Prx5 expression relative to the control after treatment with 0.3 and 0.2 mg/mL NNK (3.46-fold and 2.04-fold, respectively). Simultaneous overexpression of Prx3 and other Prx genes indicates that Prxs work synergistically to protect cells against oxidative stress. The transcription levels of Prx3 and Prx5 showed an initial
decreasing trend, followed by an increase, and then a decrease; Prx6 also showed the same expression trend. The expression levels of Prx1 and Prx3-Prx6 were assessed by Western blot and found to be very similar to their expression at the transcriptional level. The results above suggest that Prx5 is more sensitive to induction by low concentrations of NNK (0.2 mg/mL) than high concentrations. By contrast, Prx1, Prx2, Prx3, Prx4, and Prx6 are more sensitive to induction by high concentrations NNK (0.3 or 0.4 mg/mL) than low concentrations (Figure 3).
-
Table Supplementary Table 1. Characteristics of the Six Prxs from Hamster
Gene GenBank Accession
NumberPrx Type Putative Subcellular
LocalizationDeduced Number
of Amino AcidIsoelectric
PointMolecular Mass
(kD)Prx1 EGV94592 2Cys
peroxiredoxinCytosol 199 7.60 22.23 Prx2 EGW00605 2Cys
peroxiredoxinCytosol 198 5.35 21.81 Prx3 EGV96726 2Cys
peroxiredoxinCytosol, mitochondria,
and peroxisomes257 6.79 28.13 Prx4 XP_007650248 2Cys
PeroxiredoxinSecretion from the cell 274 6.40 31.15 Prx5 ERE77746 Atypical 2Cys
PeroxiredoxinMitochondria 213 9.35 22.56 Prx6 ERE72683 1Cys
PeroxiredoxinCytosol 231 6.43 25.88 Table Supplementary Table 2. The Sequences of Gene-Specific Primers for qRT-PCRs Used in This Study
Gene Forward Primers (5'–3') Reverse Primers (5'–3') Prx1 TATCCTGCTCCCAACTTCAA GTCCCAATCCTCCTTGTTTC Prx2 TGATGAGGGCATTGCTTACA TCTACAGAGCGTCCCACAGG Prx3 CCCACTTCAGTCATCTTGCC CAACAGCACTCCGTAGTCTCG Prx4 CGAAGACAAGGAGGACTGGG AGGCTTGAACCAAACGCAGT Prx5 TCAAGGGCAAGAAAGGTGTT GCCGACGATTCCCAAAGA Prx6 TTGATGATAAGGGCAGGGAC GGTGGCAGGGTAGAGGATAG GAPDH CGTGCCTGGAGAAACCTG AGAGTGGGAGTTGCTGTTGAAGTCG 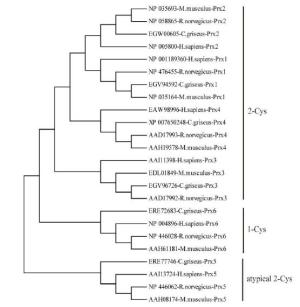
Figure Supplementary Figure S1. Neighbor-joining tree of the Prxs amino acid sequences of hamster, R.novegicus, M. muschlus and H. sapiens. Bootstrap values (100 replicates) of > 50 are shown.

Figure Supplementary Figure S2. Multiple sequence alignments of the six Prx proteins from hamster. Idential residues are shaded black, while similar residues are gray. Two conserved functional active cysteine sites and two H2O2 sensitive motifs (GGLG and YF) are highlighted in black boxes. Predicted signal peptide for Prx4 is underlined.
doi: 10.3967/bes2017.031
Characterization and Expression Analysis of Peroxiredoxin Genes in NNK-induced V79 Cells
-
-
Figure 3. Expression of (A) Prx1, (B) Prx2, (C) Prx3, (D) Prx4, (E) Prx5, and (F) Prx6 in V79 cells after NNK induction. Bars represent SD, and data points reflect the means of three independent experiments. Values above bars indicate fold-increases in inducible transcription level relative to that of the control.
Figure Supplementary Figure S2. Multiple sequence alignments of the six Prx proteins from hamster. Idential residues are shaded black, while similar residues are gray. Two conserved functional active cysteine sites and two H2O2 sensitive motifs (GGLG and YF) are highlighted in black boxes. Predicted signal peptide for Prx4 is underlined.
Table Supplementary Table 1. Characteristics of the Six Prxs from Hamster
Gene GenBank Accession
NumberPrx Type Putative Subcellular
LocalizationDeduced Number
of Amino AcidIsoelectric
PointMolecular Mass
(kD)Prx1 EGV94592 2Cys
peroxiredoxinCytosol 199 7.60 22.23 Prx2 EGW00605 2Cys
peroxiredoxinCytosol 198 5.35 21.81 Prx3 EGV96726 2Cys
peroxiredoxinCytosol, mitochondria,
and peroxisomes257 6.79 28.13 Prx4 XP_007650248 2Cys
PeroxiredoxinSecretion from the cell 274 6.40 31.15 Prx5 ERE77746 Atypical 2Cys
PeroxiredoxinMitochondria 213 9.35 22.56 Prx6 ERE72683 1Cys
PeroxiredoxinCytosol 231 6.43 25.88 Table Supplementary Table 2. The Sequences of Gene-Specific Primers for qRT-PCRs Used in This Study
Gene Forward Primers (5'–3') Reverse Primers (5'–3') Prx1 TATCCTGCTCCCAACTTCAA GTCCCAATCCTCCTTGTTTC Prx2 TGATGAGGGCATTGCTTACA TCTACAGAGCGTCCCACAGG Prx3 CCCACTTCAGTCATCTTGCC CAACAGCACTCCGTAGTCTCG Prx4 CGAAGACAAGGAGGACTGGG AGGCTTGAACCAAACGCAGT Prx5 TCAAGGGCAAGAAAGGTGTT GCCGACGATTCCCAAAGA Prx6 TTGATGATAAGGGCAGGGAC GGTGGCAGGGTAGAGGATAG GAPDH CGTGCCTGGAGAAACCTG AGAGTGGGAGTTGCTGTTGAAGTCG -
[1] Siegel R, Ward E, Brawley O, et al. The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin, 2011, 61: 212-36. doi: 10.3322/caac.v61:4 [2] Wu JJ, Yang T, Li X, et al. 4-(methylnitrosamino)-1-(3-pyridyl) -1-butanone Induces circulating microRNA deregulation in early lung carcinogenesis. Biomed Environ Sci, 2014, 27: 10-6. http://www.besjournal.com/Articles/Archive/2014/No1/201402/t20140212_93403.html [3] Li LQ, Yu AQ. The functional role of peroxiredoxin 3 in reactive oxygen species, apoptosis, and chemoresistance of cancer cells. J Cancer Res Clin Oncol, 2015, 141: 2071-7. doi: 10.1007/s00432-015-1916-3 [4] Rostila A, Puustinen A, Toljamo T, et al. Peroxiredoxins and tropomyosins as plasma biomarkers for lung cancer and asbestos exposure. Lung Cancer, 2012, 77: 450-9. doi: 10.1016/j.lungcan.2012.03.024 [5] Lehtonen ST, Svensk AM, Soini Y, et al. Peroxiredoxins, a novel protein family in lung cancer. Int J Cancer, 2004, 111: 514-21. doi: 10.1002/ijc.v111:4 [6] Rostila A, Puustinen A, Toljamo T, et al. Peroxiredoxins and tropomyosins as plasma biomarkers for lung cancer and asbes-tos exposure. Lung Cancer, 2012, 77: 450-9. doi: 10.1016/j.lungcan.2012.03.024 [7] Chen MF, Chen WC, Wu CT, et al. p53 status is a major determinant of effects of decreasing peroxiredoxin I expression on tumor growth and response of lung cancer cells to treatment. Int J Radiat Oncol Biol Phys, 2006, 66: 1461-72. doi: 10.1016/j.ijrobp.2006.07.1372 [8] Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell, 2012, 48: 158-67. doi: 10.1016/j.molcel.2012.09.025 [9] Chuang CH, Hu ML. Synergistic DNA damage and lipid peroxidation in cultured human white blood cells exposed to 4-(methyl-nitrosamino)-1-(3-pyridyl)-1-butanone and ultraviolet A. Environ Mol Mutagen, 2006, 47: 73-81. doi: 10.1002/(ISSN)1098-2280 [10] Gupta P, Ghergherehchi L, Kulp GA, et al. Effect of tobacco specific N-nitrosamines, NNN and NNK on cultured retinal pigment epithelial cells:Implications to age related macular degeneration. In Proceedings of ARVO 2011 Visionary Genomics, Fort Lauderdale, FL, USA, 1-5 May 2011. -




 下载:
下载:



 Quick Links
Quick Links