-
Diabetes mellitus, a complex and chronic disease, has become a major public health problem and imposes new challenges on health systems[1]. The International Diabetes Federation's latest assessment shows that 387 million people (8.3% of adults) suffer from diabetes, and the number of patients is expected to rise above 592 million by 2035[2]. Diabetes has had a massive impact on China in recent decades. Growing evidence shows that China has a very high incidence of diabetes, and approximately 92.4 million adults currently have diabetes in China[3]. Type 2 diabetes (T2DM), the most common form of diabetes, is characterized by insulin resistance[4]. Several studies have indicated that susceptibility genes and environmental factors play significant roles in the development of T2DM[5-6]. Multiple genes are thought to be involved, each producing a small effect on T2DM risk[7].
Despite the differing pathogenesis of type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM), epidemiological data have indicated that these two types of diabetes display familial clustering, which suggests a common genetic basis between them[8]. Several susceptibility genes have been reported to be associated with both T1DM and T2DM[9]. The small ubiquitin-like modifier (SUMO) is a protein that attaches to target proteins and controls the target proteins' stability or activity[10]. SUMO4 is the fourth member of the SUMO family and is mainly expressed in the immune system[11]. The SUMO4 gene is located at a T1DM susceptibility locus (IDDM5) on chromosome 6q25, and a single nucleotide polymorphism (SNP) is associated with susceptibility to T1DM[11-13]. In vitro studies have shown that SUMO4 negatively regulates nuclear factor κB (NF-κB) transcriptional activity, which is a key transcriptional pathway involved in immune response and inflammation[11]. Inflammation induces inhibition of the insulin signaling pathway, which leads to insulin resistance and contributes to the development of T2DM[14].
Some epidemiology studies reported that the SUMO4 M55V polymorphism is associated with susceptibility to T2DM[15-19], while other studies did not favor the association[20-23]. For example, Noso et al. (2009) reported that the SUMO4 M55V polymorphism is associated with an increased risk of T2DM in a population recruited from the western region of Japan[15], whereas other studies conducted in Wakayama and Toyoto did not confirm this association, neither in an allelic model, nor in additive, dominant, and recessive models[20]. Therefore, the association between the SUMO4 M55V polymorphism and T2DM remained controversial. Two separate meta-analyses were conducted to explore the association between the SUMO4 M55V polymorphism and T2DM, and revealed an association[24-25]. However, these studies only conducted meta-analyses based on alleles or combined genotypes, without taking genetic models into account.
The patterns of inheritance of the SUMO4 M55V polymorphism are not clear, and more than half of the studies did not support the association between the SUMO4 M55V polymorphism and T2DM. Therefore, we preformed this updated meta-analysis to validate the association between the SUMO4 M55V polymorphism and T2DM under dominant, recessive, co-dominant (homozygous and heterozygous), and additive models.
-
A comprehensive literature search was conducted of the online databases MEDLINE (Medical Literature Analysis and Retrieval System Online), China National Knowledge Infrastructure (CNKI), Sinomed, and WanFang up to December 21, 2016. The following medical subject headings and keywords were used in the search strategy: 'Small ubiquitin-like modifier 4' or 'SUMO4, ' 'type 2 diabetes mellitus' or 'type 2 diabetes' or 'T2DM, ' and 'rs237025' or 'Met55Val' or 'M55V, ' or 'A163G.' References lists of the retrieved articles and reviews were also screened for additional articles not captured by electronic search.
Eligible studies included in the meta-analysis met all the following criteria: (1) the associations of polymorphisms in the SUMO4 gene with T2DM; (2) case-control or cohort design; (3) provided odds ratio (OR) with 95% confidence interval (CI) or genotype frequency among case and control group; (4) study samples being unrelated individuals drawn from clearly defined populations; (5) written in English or Chinese; (6) for duplicate publications from the same population, only the paper that had the largest population, contained more useful information, or the latest one was selected. The exclusion criteria were defined as studies on animals, case reports, reviews, abstracts, editorial comments, and reports with incomplete data.
-
The following information was extracted: first author, year of publication, ethnicity, country of origin, sample size, age, gender, allele/genotypic frequencies and minor allele frequency in cases and controls and P-value for the allele frequency. The quality of studies was evaluated independently by two investigators (ZHANG Qun and LIU Di) according to the Newcastle-Ottawa Scale (NOS). Uncertainties were resolved by discussions or by consensus with a third reviewer (ZHAO Zhong Yao). NOS evaluated studies with a star-rating system ranging from 0 (lowest) to 8 (highest) stars, which was based on three study components including selection, comparability, and outcome assessment. Studies with more than 5 points were evaluated as qualified.
-
The associations of polymorphisms in the SUMO4 gene with T2DM were estimated by calculating pooled OR and 95% CI by using RevMan 5.3. Hardy-Weinberg equilibrium (HWE) among controls was evaluated by chi-square test and P < 0.05 was considered as significant disequilibrium. The chi-square test and the inconsistency index (I2) were applied to assess heterogeneity among studies. The Z-test was used to calculate the P-value of the overall effect for the meta-analysis. Because the true effect size might differ from study to study, random effects models were used for combined data, regardless of whether or not heterogeneity was detected in the meta-analysis. Pooled OR and 95% CI were computed by the random-effects method of Mantel-Haenszel for combined data. Publication bias was evaluated by Begg's funnel plot and Egger's test. All the tests were two-sided, and results were considered statistically significant at P ≤ 0.05 unless otherwise stated.
-
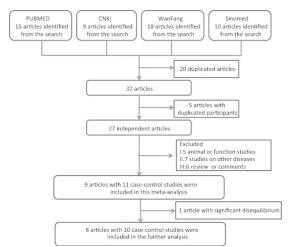
A total of 52 articles were identified after an initial search, 20 of which were duplicate articles (Figure 1). Twenty-three articles were further excluded because of duplicate sample use (5), animal or function studies (4), studies on other diseases (8), and review or comments (6) (Figure 1). From the above, we identified nine articles, which were included in the SUMO4 M55V analysis (Table 1). One study was further excluded because of significant disequilibrium in the Hardy-Weinberg equilibrium test. Finally, eight articles including 10 case-control studies, with a total of 2932 cases and 2679 controls, were included in this meta-analysis. The study quality is summarized in Table 2. The quality of the studies included in this meta-analysis was acceptable (at least 5 points).
Table 1. Characteristics of Case-control Studies Included in Meta-analysis
Study Year Ethnicity/Region Population Group Subject Size Diagnosis Genotyping Method P-value for HWE Noso S[15] 2007 Western area Japanese T2DM 355 ADA Taqman SNP genotyping assay 0.690 Control 398 0.291 Shimada T[20] 2009 Wakayama Japanese T2DM 423 WHO Taqman SNP genotyping assay 0.716 Control 436 0.094 Shimada T[20] 2009 Tokyo Japanese T2DM 451 WHO 0.408 Control 469 0.378 Ji Z[17] 2010 Hunan Chinese T2DM 427 WHO PCR-RFLP 0.830 Control 281 0.079 Li B[21] 2011 Yunan Chinese T2DM 232 WHO PCR-RFLP 0.260 Control 102 0.126 Lin HY[16] 2007 Taiwan Chinese T2DM 574 NA PCR-RFLP 0.193 Control 323 0.466 Fallah S[22] 2010 Tehran Irani T2DM 50 NA PCR-RFLP 0.493 Control 50 0.998 Hu RT[23] 2009 Va Chinese T2DM 96 WHO PCR-RFLP 0.487 Control 104 0.673 Hu RT[23] 2009 Lalu Chinese T2DM 54 WHO PCR-RFLP 0.088 Control 234 0.817 Pu LM[18] 2012 Beijing Chinese T2DM 270 WHO PCR-HRM 1.92 × 10-5 Control 282 0.086 Note. ADA: American Diabetes Association. WHO: World Health Organization. PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism restriction fragment length polymorphism. NA: not reported in the paper. Table 2. Assessment of Case-control Studies Using Newcastle-Ottawa Scale for Evaluating Methodological Quality
Study Case Definition Representativeness of the Case Selection of Controls Definition of Controls Comparability Ascertainment of Exposure Same Method of Ascertainment for Cases and Controls Non-Response Rate Noso S 2007[15] √ √ √ √ √ √ √ √ Shimada T 2009[20] √ √ √ √ √ √ √ Shimada T 2009[20] √ √ √ √ √ √ √ Lin HY 2007[16] √ √ √ √ √ √ √ Ji Z 2010[17] √ √ √ √ √ √ Li B 2011[21] √ √ √ √ √ √ √ Hu RT 2009[23] √ √ √ √ √ √ √ Hu RT 2009[23] √ √ √ √ √ √ √ Pu LM 2012[18] √ √ √ √ √ √ √ √ Fallah S 2010[12] √ √ √ √ √ √ -
Figure 2 presents the forest plot of the association between the SUMO4 M55V polymorphism (also named G163A in SUMO4, or rs237025) and T2DM in each studies and the meta analysis. The significant association between the SUMO4 M55V polymorphism and susceptibility to T2DM was observed in the dominant model (GG + GA versus AA: OR = 1.21, 95% CI = 1.05-1.40, P = 0.009), recessive model (GG versus GA + AA: OR = 1.29, 95% CI = 1.06-1.56, P = 0.010), homozygous model (GG versus AA: OR = 1.41, 95% CI = 1.15-1.71, P = 0.001), and additive model (G versus A: OR = 1.18, 95% CI = 1.08-1.29, P= 0.001), and marginally significant in the heterozygous model (GA versus AA: OR = 1.16, 95%CI = 0.98-1.36, P = 0.080).
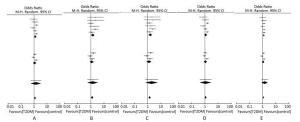
Figure 2. Forest plots of meta-analysis of the association between SUMO4 M55V polymorphisms and T2DM in a dominant model (A), recessive model (B), homozygous model (C), heterozygous model (D), and additive model (E).
No significant subgroup differences were observed (P ranged from 0.52 to 0.56 in all five genetic models). Subgroup analysis showed significant associations between the SUMO4 M55V polymorphism and susceptibility to T2DM in the Chinese population under all but the heterozygous model (GA versus AA: OR = 1.12, 95% CI = 0.86-1.44, P = 0.090) and marginally significant associations in the Japanese population (P ranged from 0.13 to 0.40 in all five genetic models).
-
Funnel plot asymmetry was evaluated by Egger's regression test. If the line passed through the origin, it would indicate the absence of publication bias. The funnel plots and Egger's linear regression test are shown in Figure 3. No publication biases were detected under the five genetic models (all P > 0.05).
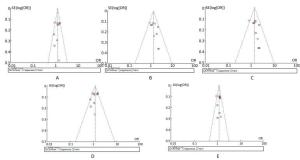
Figure 3. Publication biases indicated by the funnel plots. Figure A, dominant model, P = 0.655 and 0.550 in Begg's and Egger's test, respectively. Figure B, recessive model, P = 0.245 and 0.063 in Begg's and Egger's test, respectively. Figure C, homozygous model, P = 0.074 and 0.131 in Begg's and Egger's test, respectively. Figure D, heterozygous model, P = 0.655 and 0.237 in Begg's and Egger's test, respectively. Figure E, additive model, P = 0.788 and 0.866 in Begg's and Egger's test, respectively.
A sensitivity analysis was conducted to explore the sources of heterogeneity, and the results showed that no single study affected the pooled OR and CIs in a qualitative manner.
-
Our study found a statistically significant association between the SUMO4 M55V polymorphism and T2DM under dominant, recessive, co-dominant (homozygous and heterozygous), and additive models, especially in the Chinese population. In addition, publication bias tests and sensitivity analysis showed that the overall results were robust. The implication of this finding is that subjects with the G allele in the SUMO4 M55V polymorphism are at a high risk of developing T2DM late in life.
The meta-analysis demonstrated that the SUMO4 M55V polymorphism was associated with T2DM, consistent with several domestic and foreign reports[15-19]. In subjects living in the western region of Japan, Noso et al. (2007)[15] found that the frequency of the G allele was higher in T2DM patients (P < 0.05), and the frequency of the GG and GA genotypes in the case group was also higher than in controls, whereas these findings were not validated in subjects recruited from Wakayama and Toyoto of Japan[20]. A study in the Taiwan, China population showed that SUMO4 M55V polymorphism G allele carriers have a higher risk of suffering from T2DM[16], as did two studies conducted in Beijing and Hubei[17-18]. However, studies conducted in Han Chinese and two ethnicities (Va, Lahu) recruited from the Yunan province of China did not demons-trate the same association[21-23]. In addition, Fallah et al. (2010) found that the SUMO4 M55V polymorphism was not associated with T2DM in the Iranian population[22], while Sozen et al. (2014) reported that the SUMO4 M55V polymorphism was associated with T2DM in the Turkish population[19]. The study conducted in Turkey was excluded from this meta-analysis because of the significant Hardy-Weinberg disequilibrium, while the study conducted in Iran may have lacked statistical power (50 in each of disease and control groups). This disparity in the association between the SUMO4 M55V polymorphism and T2DM between studies might be caused by heterogeneity in ethnicity and genetic background[26]. In addition, the relatively small sample size in several studies also contributes to this discrepancy.
Genome-wide scans for T2DM revealed linkage of T2DM at chromosome 6q, where a susceptibility gene for T2DM (1DDM5) was mapped in African-American[27], Chinese[28], and Finnish populations[29]. In addition, strong associations between the SUMO4 M55V polymorphism and T2DM were validated in several meta-analyses[24, 30]. The consistency in the association between the SUMO4 M55V polymorphism and T1DM or T2DM suggested that SUMO4 may explain the common genetic predisposition to T1DM and T2DM.
SUMO4 regulates the activation of IκBα by sumoylation, and thus negatively regulates the activity of NFκB[31]. IκBα inhibits NF-κB by masking the nuclear localization signals (NLS) of NF-κB proteins and keeping them sequestered in an inactive state in the cytoplasm; it also blocks the ability of NF-κB transcription factors to bind to DNA, which is required for proper functioning of NF-κB[32]. NF-κB, a transcriptional factor for inflammation, is relevant to the pathogenesis of diabetes mellitus[33]. In addition, the interaction between NFκB and IκBα result in the insulin resistance in vivo and in vitro studies[34-37]. Furthermore, the association of SUMO4 Met55Val variation with increased insulin resistance was reported in newly diagnosed T2DM in a Chinese population[38]. Therefore, the Met55Val variation in SUMO4 may affect its capacity to modify IκBα, leading to increased activity of NFκB and increased insulin resistance and thus increase the risk of diabetes mellitus.
However, there are two limitations to our meta-analysis. First, confounding factors were not accounted for because most of the studies did not consider these. Unmeasured confounding factors might affect the observed association. Secondly, heterogeneity might lead to publication bias, because studies with negative results might not be published. Although the funnel plots and Egger's linear regression test indicated no biases, it is possible hat the relatively small number of studies included in this meta-analysis led to less statistical power to detect publication biases.
To summarize, our meta-analysis demonstrated that the G allele in the SUMO4 M55V polymorphism may be a susceptible risk locus to T2DM, especially in the Chinese population. The disparity among studies suggests that further investigations are required on the associations between the SUMO4 M55V polymorphism and T2DM susceptibility in different ethnic groups. The SUMO4 M55V polymorphism might be a causal factor in T2DM via its regulation of NFκB signaling.
doi: 10.3967/bes2017.038
Association between the SUMO4 M55V Polymorphism and Susceptibility to Type 2 Diabetes Mellitus: A Meta-analysis
-
Abstract:
Objective The aim of this study is to determine whether the SUMO4 M55V polymorphism is associated with susceptibility to type 2 diabetes mellitus (T2DM). Methods A meta-analysis was performed to detect the potential association of the SUMO4 M55V polymorphism and susceptibility to T2DM under dominant, recessive, co-dominant (homogeneous and heterogeneous), and additive models. Results A total of eight articles including 10 case-control studies, with a total of 2932 cases and 2679 controls, were included in this meta-analysis. The significant association between the SUMO4 M55V polymorphism and susceptibility to T2DM was observed in the dominant model (GG + GA versus AA: OR = 1.21, 95% CI = 1.05-1.40, P= 0.009), recessive model (GG versus GA + AA: OR = 1.29, 95% CI = 1.07-1.356, P= 0.010), homozygous model (GG versus AA: OR = 1.41, 95% CI = 1.06-1.56, P= 0.001), and additive model (G versus A: OR = 1.18, 95% CI = 1.08-1.29, P=0.001), and marginally significant in the heterozygous model (GA versus AA: OR = 1.16, 95% CI = 0.98-1.36, P= 0.080). In subgroup analyses, significant associations were observed in the Chinese population under four genetic models excluding the heterozygous model, whereas no statistically significant associations were observed in the Japanese population under each of the five genetic models. Conclusion The meta-analysis demonstrated that the G allele of the SUMO4 M55V polymorphism could be a susceptible risk locus to T2DM, mainly in the Chinese population, while the association in other ethnic population needs to be further validated in studies with relatively large samples. -
Key words:
- SUMO4 /
- Type 2 diabetes mellitus (T2DM) /
- Polymorphisms /
- Meta-analysis
注释:1) CONFLICT OF INTEREST: 2) AUTHORS CONTRIBUTIONS: -
Figure 3. Publication biases indicated by the funnel plots. Figure A, dominant model, P = 0.655 and 0.550 in Begg's and Egger's test, respectively. Figure B, recessive model, P = 0.245 and 0.063 in Begg's and Egger's test, respectively. Figure C, homozygous model, P = 0.074 and 0.131 in Begg's and Egger's test, respectively. Figure D, heterozygous model, P = 0.655 and 0.237 in Begg's and Egger's test, respectively. Figure E, additive model, P = 0.788 and 0.866 in Begg's and Egger's test, respectively.
Table 1. Characteristics of Case-control Studies Included in Meta-analysis
Study Year Ethnicity/Region Population Group Subject Size Diagnosis Genotyping Method P-value for HWE Noso S[15] 2007 Western area Japanese T2DM 355 ADA Taqman SNP genotyping assay 0.690 Control 398 0.291 Shimada T[20] 2009 Wakayama Japanese T2DM 423 WHO Taqman SNP genotyping assay 0.716 Control 436 0.094 Shimada T[20] 2009 Tokyo Japanese T2DM 451 WHO 0.408 Control 469 0.378 Ji Z[17] 2010 Hunan Chinese T2DM 427 WHO PCR-RFLP 0.830 Control 281 0.079 Li B[21] 2011 Yunan Chinese T2DM 232 WHO PCR-RFLP 0.260 Control 102 0.126 Lin HY[16] 2007 Taiwan Chinese T2DM 574 NA PCR-RFLP 0.193 Control 323 0.466 Fallah S[22] 2010 Tehran Irani T2DM 50 NA PCR-RFLP 0.493 Control 50 0.998 Hu RT[23] 2009 Va Chinese T2DM 96 WHO PCR-RFLP 0.487 Control 104 0.673 Hu RT[23] 2009 Lalu Chinese T2DM 54 WHO PCR-RFLP 0.088 Control 234 0.817 Pu LM[18] 2012 Beijing Chinese T2DM 270 WHO PCR-HRM 1.92 × 10-5 Control 282 0.086 Note. ADA: American Diabetes Association. WHO: World Health Organization. PCR-RFLP: Polymerase Chain Reaction-Restriction Fragment Length Polymorphism restriction fragment length polymorphism. NA: not reported in the paper. Table 2. Assessment of Case-control Studies Using Newcastle-Ottawa Scale for Evaluating Methodological Quality
Study Case Definition Representativeness of the Case Selection of Controls Definition of Controls Comparability Ascertainment of Exposure Same Method of Ascertainment for Cases and Controls Non-Response Rate Noso S 2007[15] √ √ √ √ √ √ √ √ Shimada T 2009[20] √ √ √ √ √ √ √ Shimada T 2009[20] √ √ √ √ √ √ √ Lin HY 2007[16] √ √ √ √ √ √ √ Ji Z 2010[17] √ √ √ √ √ √ Li B 2011[21] √ √ √ √ √ √ √ Hu RT 2009[23] √ √ √ √ √ √ √ Hu RT 2009[23] √ √ √ √ √ √ √ Pu LM 2012[18] √ √ √ √ √ √ √ √ Fallah S 2010[12] √ √ √ √ √ √ -
[1] Canivell S, Gomis R. Diagnosis and classification of autoimmune diabetes mellitus. Autoimmun Rev, 2014; 13, 403-7. doi: 10.1016/j.autrev.2014.01.020 [2] International Diabetes Federation. Annual Report 2013-2014. http://www.idf.org/regions/EUR/annualreports. [2016-12-26] [3] Yang W, Lu J, Weng J, et al. Prevalence of diabetes among men and women in China. N Engl J Med, 2010; 362, 1090-101. doi: 10.1056/NEJMoa0908292 [4] Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell, 2012; 14, 1160-71. [5] Hivert MF, Vassy JL, Meigs JB. Susceptibility to type 2 diabetes mellitus-from genes to prevention. Nat Rev Endocrinol, 2014; 10, 198-205. doi: 10.1038/nrendo.2014.11 [6] Hanson RL, Muller YL, Kobes S, et al. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes, 2014; 63, 369-76. doi: 10.2337/db13-0416 [7] Ahlqvist E, Ahluwalia TS, Groop L. Genetics of type 2 diabetes. Clin Chem, 2011, 57; 241-54. doi: 10.1373/clinchem.2010.157016 [8] Li H, Lindholm E, Almgren P, et al. Possible human leukocyte antigen-mediated genetic interaction between type 1 and type 2 Diabetes. J Clin Endocrinol Metab, 2001; 86, 574-82. https://www.researchgate.net/profile/Eero_Lindholm/publication/12171696_Possible_Human_Leukocyte_Antigen-Mediated_Genetic_Interaction_between_Type_1_and_Type_2_Diabetes_1/links/54ddb7180cf25b09b91484b1/Possible-Human-Leukocyte-Antigen-Mediated-Genetic-Interaction-between-Type-1-and-Type-2-Diabetes-1.pdf [9] Eftychi C, Howson JM, Barratt BJ, et al. Analysis of the type 2 diabetes-associated single nucleotide polymorphisms in the genes IRS1, KCNJ11, and PPARG2 in type 1 diabetes. Diabetes, 2004; 53, 870-3. doi: 10.2337/diabetes.53.3.870 [10] Kroetz MB. SUMO: a ubiquitin-like protein modifier. Yale J Biol Med, 2005; 78, 197-201. http://europepmc.org/articles/PMC2259148?pdf=render [11] Guo D, Li M, Zhang Y, et al. A functional variant of SUMO4, a new Ⅰ kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet, 2004; 36, 837-41. doi: 10.1038/ng1391 [12] Tsurumaru M, Kawasaki E, Ida H, et al. Evidence for the role of small ubiquitin-like modifier 4 as a general autoimmunity locus in the Japanese population. J Clin Endocrinol Metab, 2006; 91, 3138-43. doi: 10.1210/jc.2006-0206 [13] Bohren KM, Nadkarni V, Song JH, et al. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type Ⅰ diabetes mellitus. J Biol Chem, 2004; 279, 27233-8. doi: 10.1074/jbc.M402273200 [14] Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol, 2011; 11, 98-107. doi: 10.1038/nri2925 [15] Noso S, Fujisawa T, Kawabata Y, et al. Association of small ubiquitin-like modifier 4 (SUMO4) variant, located in IDDM5 locus, with type 2 diabetes in the Japanese population. J Clin Endocrinol Metab, 2007; 92, 2358-62. doi: 10.1210/jc.2007-0031 [16] Lin HY, Wang CL, Hsiao PJ, et al. SUMO4 M55V variant is associated with diabetic nephropathy in type 2 diabetes. Diabetes, 2007; 56, 1177-80. doi: 10.2337/db06-1283 [17] Ji Z, Dai Z, Xu Y. Association between small ubiquitin-like modifer 4 M55V polymorphism with type 2 diabetes and related factors. Chin J Diabetes Mellitus, 2010; 2, 344-8. (In Chinese) [18] Pu LM, Nan N, Yang ZN, et al. Association between SUMO4 polymorphisms and type 2 diabetes mellitus. Hereditas (Beijing), 2012; 3, 315-25. (In Chinese) [19] Sozen S, Horozoglu C, Bireller ES, et al. Association of SUMO4 M55V and-94ins/del gene variants with type-2 diabetes. In Vivo, 2014; 28, 919-23. http://iv.iiarjournals.org/content/28/5/919.full [20] Shimada T, Furukawa Y, Furuta H, et al. SUMO4 Met55Val polymorphism is associated with coronary heart disease in Japanese type 2 diabetes individuals. Diabetes Res Clin Pract, 2009; 85, 85-9. doi: 10.1016/j.diabres.2009.04.001 [21] Li B, Li HB, Wang YM, et al. The association of SUMO4 Gene 163 A/G polymorphism with type 2 diabetic nephropathy. Chin J Diabetes, 2011; 19, 726-8. (In Chinese) http://en.cnki.com.cn/Article_en/CJFDTOTAL-ZGTL201110004.htm [22] Fallah S, Jafarzadeh M, Hedayati M. No association of the SUMO4 polymorphism M55V variant in type 2 diabetes in Iranian subjects. Diabetes Res Clin Pract, 2010; 90, 191-5. doi: 10.1016/j.diabres.2010.05.033 [23] Hu RT, Song DP. The association of SUMO4 Gene 163 A/G polymorphism and dysglycemia in Va and Lalu population. Kunming Medical University, 2009. (In Chinese) [24] Tang ST, Peng WJ, Wang CJ, et al. Polymorphism M55V in gene encoding small ubiquitin-like modifier 4 (SUMO4) protein associates with susceptibility to type 1 (and type 2) diabetes. Diabetes Metab Res Rev, 2012; 28, 679-87. doi: 10.1002/dmrr.v28.8 [25] Meng QJ, Wang JS, Jiang J, et al. A meta-analysis on association between SUMO4 M55V polymorphism and type 2 diabetes mellitus. Chin J Diabetes, 2013; 21, 591-3. [26] Khodaeian M, Enayati S, Tabatabaei-Malazy O, et al. Association between Genetic Variants and Diabetes Mellitus in Iranian Populations: A Systematic Review of Observational Studies. J Diabetes Res, 2015; 2015, 585917. https://www.researchgate.net/publication/274631365_Association_between_Genetic_Variants_and_Diabetes_Mellitus_in_Iranian_Populations_A_Systematic_Review_of_Observational_Studies [27] Sale MM, Freedman BI, Langefeld CD, et al. A genome-wide scan for type 2 diabetes in African-American families reveals evidence for a locus on chromosome 6q. Diabetes, 2004; 53, 830-7. doi: 10.2337/diabetes.53.3.830 [28] Xiang K, Wang Y, Zheng T, et al. Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21-q23 and chromosome 1q21-q24. Diabetes, 2004; 53, 228-34. doi: 10.2337/diabetes.53.1.228 [29] Silander K, Scott LJ, Valle TT, et al. A large set of Finnish affected sibling pair families with type 2 diabetes suggests susceptibility loci on chromosomes 6, 11, and 14. Diabetes, 2004; 53, 821-9. doi: 10.2337/diabetes.53.3.821 [30] Song GG, Choi SJ, Ji JD, et al. Association between the SUMO4 M55V (A163G) polymorphism and susceptibility to type 1 diabetes: A meta-analysis. Human Immunology, 2012; 73, 1055-9. doi: 10.1016/j.humimm.2012.07.341 [31] Guo D, Li M, Zhang Y, et al. A functional variant of SUMO4, a new Ⅰ kappa B alpha modifier, is associated with type 1 diabetes. Nature Genetics, 2004; 36, 837-41. doi: 10.1038/ng1391 [32] Wang CY, Yang P, Li M, et al. Characterization of a negative feedback network between SUMO4 expression and NFkappaB transcriptional activity. Biochem Biophys Res Commun, 2009; 381, 477-81. doi: 10.1016/j.bbrc.2009.02.060 [33] Benzler J, Ganjam GK, Pretz D, et al. Central inhibition of IKKβ/NF-κB signaling attenuates high-fat diet-induced obesity and glucose intolerance. Diabetes, 2015; 64, 2015-27. doi: 10.2337/db14-0093 [34] Cai D, Yuan M, Frantz DF, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappa B. Nat Med, 2005; 11, 183-90. doi: 10.1038/nm1166 [35] Cyphert TJ, Morris RT, House LM, et al. NF-κB-dependent airway inflammation triggers systemic insulin resistance. Am J Physiol Regul Integr Comp Physiol, 2015; 309, R1144-52. doi: 10.1152/ajpregu.00442.2014 [36] Liu Z, Dou W, Ni Z, et al. Deletion of Nrf2 leads to hepatic insulin resistance via the activation of NF-κB in mice fed a high-fat diet. Mol Med Rep, 2016; 1323-31. [37] Feng H, Su R, Song Y, et al. Positive Correlation between Enhanced Expression of TLR4/MyD88/NF-κB with Insulin Resistance in Placentae of Gestational Diabetes Mellitus. PLoS One, 2016; 11, e0157185. doi: 10.1371/journal.pone.0157185 [38] Ji Z, Dai Z, Huang Y, et al. Association of SUMO4 Met55Val variation with increased insulin resistance in newly diagnosed type 2 diabetes in a Chinese population. J Huazhong Univ Sci Technolog Med Sci, 2011; 31, 306-11. doi: 10.1007/s11596-011-0372-9 -




 下载:
下载:



 Quick Links
Quick Links