-
Di (2-ethylhexyl) phthalate (DEHP) is one of the most common endocrine disruptor substances (EDS). DEHP is a plasticizer, which is most frequently detected in a variety of products and applications such as PVC interior surface coverings, food packaging, and plastic goods[1]. Human exposure to DEHP mainly occurs through ingestion of food and water[2]. Previous studies have shown that fetal exposure to DEHP may impair the endocrine function of adrenal glands in adult males and lead to reduced steroid secretion from follicular cells[3]. In recent years, studies have shown that environmental DEHP and monoethylhexyl phthalic acid (MEHP) affected lipid metabolism by activating peroxisome proliferator- activated receptor γ (PPARγ), and further led to obesity[4, 5]. Obesity disease and other abnormal metabolic diseases have been reported to occur in humans exposed to high concentrations of DEHP[5, 6]. Past studies have shown that autophagy activity in adipose tissue was significantly elevated in obese humans, and that the level of autophagy markers was significantly correlated with degree of obesity[7]. Kosacka et al., found that levels of the autophagy protein LC3 were significantly increased in the adipose tissue of type 2 diabetes patients and obese individuals[8]. As well, previous studies have shown that foods high in fat are contaminated by higher weight phthalates that are more lipophilic, such as DEHP, and that a high-fat diet (HD) and DEHP may exert a combined deleterious effect on lipid metabolism disorder[9, 10].
Studies have shown that JAK-STAT pathway is involved in lipid metabolism by either regulating expression of PPARγ, a key factor in adipogenesis and a vital target of STATs[11, 12], or directly regulating lipid metabolism-related factors such as lipoprotein lipase (LPL), the rate-limiting enzyme that promotes hydrolysis of triglycerides into free fatty acids in serum for uptake and storage by adipose tissue. JAK-STAT may also regulate the expression of peroxisomal acyl-CoA oxidase 1 (AOX), a rate-limiting enzyme in peroxisomal fatty acid oxidation which is associated with fatty acid metabolism, and fatty acid synthase (FAS), the key enzyme in de novo lipogenesis, which promotes the synthesis of long-chain fatty acids. Previous studies have indicated that TYK2 and STAT1 regulate adipogenesis and adipolysis[13, 14]. Therefore, DEHP may regulate the expression of TYK2 and STAT1 and thus affect the process of lipid metabolism. DEHP also can activate autophagy. Human umbilical vein endothelial cells exposed to MEHP exhibited significantly increased levels of autophagy, and permeability of the lysosomal and mitochondrial membranes were also significantly increased[15]. Autophagy may further increase the body's inflammatory response level and worsen insulin resistance, leading to lipid metabolic disorder[16].
However, the effect of TYK2/STAT1 and autophagy on lipid metabolic disorder induced by DEHP is unclear. We used 21-day adolescent rats to investigate the toxic effects of DEHP on lipid metabolism. We hypothesized that DEHP induced in lipid metabolism disorder through TYK2/STAT1 and autophagy.
-
All experimental rats and feed were provided by Beijing Huayu Kang Co., Ltd (Beijing, China). The high-fat feed contained: 22.3% protein, 28.5% fat, 37.14% carbohydrates, and 22.0% others (mineral, salt, vitamin, and ash content); total calories were 17.36 kJ. Normal feed contained: 19.2% protein, 8.1% fat (w/v), 54.3%, carbohydrates, and 18.4% others (mineral, salt, vitamin and ash content); total calories were 14.0 kJ. The experimental protocol was approved by the Animal Use and Care Committee of Jilin University (Changchun, China) prior to the study and is accordance with the National Institutes of Health guide for the care and use of laboratory animals. DEHP [Sinopharm Chemical Reagent Co., Shanghai, China, purity > 99% (v/v)] was dissolved in corn oil (Luhua, Shanghai, China) to 0, 2.56 × 10-3, 2.56 × 10-2, and 2.56 × 10-1 mol/L [equivalent to 0, 5, 50, 500 mg/(kg·d) exposure in rats].
A total of 160 Wistar weaning rats (age 21 days, 55 ± 5 g, 80 males and 80 females) were acclimated for one week and then randomly divided into normal diet (ND) group and high-fat diet (HD) group. Rats in each diet group were randomly divided into 4 dose groups (10 in each group) consisting of different DEHP concentrations. Doses were administered by gavage for 8 weeks. A previous survey indicated DEHP exposure of up to 4 mg/(kg·d) in some hemodialysis and transfusion patients[22]. The lowest dose setting was based on this survey. Additionally, as the dosage selection should include 100 times the dose of human exposure in subchronic and chronic toxicology experiments, doses were set in 10 fold increments to a maximum concentration of 500 mg/(kg·d) in the highest dosage. As such, DEHP exposure concentrations were 0, 5, 50, 500 mg/(kg·d). Rats were provided with ad libitum access to food and water at a temperature of 20 ± 2 ℃ and a humidity of 50% ± 20% (w/v).
-
All rats were weighed and anesthetized with 3.5% (v/v) chloral hydrate after 24 h fasting. Blood was drawn from the heart of each rat via vacuum blood collection tubes and was centrifuged at 5, 000 rpm/min for 10 min after stratification for 1 h at 4 ℃. Liver and adipose tissues were removed by laparotomy and were stored in liquid nitrogen.
-
Levels of triglyceride (TG), total cholesterol (TC), low density lipoprotein (LDL), high density lipoprotein (HDL), leptin (LEP), and adiponectin (ADP) in serum, as well as the levels of TG, and TC in liver and adipose were assayed by ELISA kit (R & D Systems, USA), in strict accordance with the manufacturer's instructions.
-
The total RNA was extracted by Trizol (TaKaRa, Kusatsu) reagent. 500 ng RNA was reverse transcribed into cDNA, SYBR Premix Ex TaqII (TaKaRa) was used according to the manufacturer's instructions, and the PCR reaction was carried out in 45 cycles of 95 ℃ for 20 s and 60 ℃ for 20 s. Tyk2, Stat1, Pparγ, Fas, Aox, Lpl, and LC3 were selected as candidate genes and β-actin was used as the internal reference. Agilent Mx3000P real-time PCR system (Japan) was used to conduct PCR. The primer sequences used for amplification of each gene are shown (Table 1).
Table 1. The Primers of Real Time Quantitative PCR
Gene Name Primer Sequence Tyk2 Forward 5'-ACTATGCATCTGATGTCTGGTCCTT-3' Reverse 5'-CAGTCATCTGGCCCTGGGTTA-3' Stat1 Forward 5'-TGGCAGCTGAGTTCCGACA-3' Reverse 5'-CTGGCACAACTGGGTTTCAAAG-3' Pparγ Forward 5'-GGAGCCTAAGTTTGAGTTTGCTGTG-3' Reverse 5'-TGCAGCAGGTTGTCTTGGATG-3' Fas Forward 5'-TGGTCACAGACGATGACAGGA-3' Reverse 5'-AGGCGTCGAACTTGGACAGA-3' Aox Forward 5'-ATTGGCACCTACGCCCAGAC-3' Reverse 5'-CCAGGCCACCACTTAATGGAA-3' Lpl Forward 5'-CATTGGAGAAGCCATTCGTGT-3' Reverse 5'-TGCCTTGCTGGGGTTTTC-3' LC3 Forward 5'-CGTCCTGGACAAGACCAAGT-3' Reverse 5'-TCTCGCTCTCGTACACTTCA-3' -
Tissues were soaked in 10% (v/v) formalin, and embedded in paraffin, which was subsequently cut into 5 μm sections using a microtome (LEICA RM2145, Germany). Sections were sequentially immersed in xylene and gradually increasing concentrations of alcohol for dewaxing. For HE staining, the dewaxed sections were dyed in hematoxylin for several minutes. Sections were rinsed with running water, placed in acid and ammonia for color separation, and washed with distilled water. Slices were subsequently placed in 70% (v/v) and 90% (v/v) alcohol, respectively, to dehydrate for 10 min. Then sections were stained with eosin for 2-3 min, dehydrated with pure alcohol, sections were then cleared in xylene and mounted in gum. For IHC, sections were immersed into 10% (v/v) H2O2 for 10 min to dispel endogenous catalase and then a 10-fold dilution of rat serum was applied to block nonspecific binding. Next, liver tissue sections were incubated with antibodies (as shown in Western blot below). Each section was incubated with the appropriate secondary antibody for 30 min after washing 3 times for 5 min using 1× PBS, and finally incubated with streptomycin-peroxidase complex at room temperature for 10 min. Protein levels were detected by Image-pro plus software.
-
Proteins were extracted from adipose and liver tissues by RIPA lysis buffer, and concentration was determined by bicinchoninic acid protein assay kit (Beyotime). Equal amounts of protein were subjected to 10% SDS-PAGE prior to transfer onto PVDF membranes (Millipore). After blocking with 5% nonfat milk, the membranes were incubated with antibody including anti-mouse TYK2 (Santa Cruz Biotechnology, sc-5271, 1:500), anti-rabbit STAT1 (Proteintech, 10144-2-AP, 1:500), anti-rabbit PPARγ (Abcam, ab191407, 1:1, 000), anti-rabbit AOX (Proteintech, 10957-1-AP, 1:500), anti-rabbit FAS (Proteintech, 10624-2-AP, 1:500), anti-mouse LPL (Abcam, ab93898, 1:1, 000), anti-mouse LC3 (Santa Cruz, sc-398822, 1:500), anti-mouse β-actin (Proteintech, 60008-1-lg, 1:2, 000), rabbit anti-phospho-TYK2 (Arigo, AGR51647, 1:500), and rabbit anti-phospho-STAT1 (Cell Signaling, #7649, 1:1, 000). The internal reference protein was mouse β-actin. Membranes were subsequently washed and incubated with goat anti-rabbit IgG (Proteintech, SA00001-2, 1:2, 000) or the goat anti-mouse IgG (Proteintech, SA00001-1, 1:2, 000) secondary antibodies. After secondary antibody incubation and prior to imaging, enhanced chemiluminescence substrate (Beyotime) was applied for 5 min. Relative protein levels were quantified via densiometric scan and Image J.
-
SPSS 18.0 statistical software (SPSS Inc. Chicago, Illinois, USA) was used for statistical analysis. According to normality of variance, the data were listed as mean ± standard deviation. One-way ANOVA followed by Tukey-Kramer post-hoc test for multiple comparisons was utilized to test for differences among groups. P < 0.05 was considered statistically significant.
-
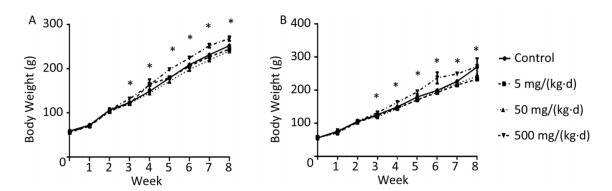
The body weight of rats on both diets increased with exposure time (Figure 1A, B). As differences were not found in body weight, lipids, or hormones between male and female rats, the sex was combined for analysis in the following results (Supplementary Figure S1, Supplementary Tables S1-S2 available in www.besjournal.com). The body weight of ND rats was significantly higher in only the 500 mg/(kg·d) dose group when compared to the other three dose groups (P < 0.05). Similar to the ND treatment, body weight in 500 mg/(kg·d) group was higher when compared with other groups in HD (P < 0.05), while the weight in 5 mg/(kg·d) group was lower than both the control and 50 mg/(kg·d) groups (P < 0.05). There was no significant difference between ND and HD treatments.
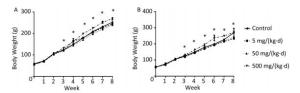
Figure 1. Effects of DEHP and HD on body weight of rats. (A) Body weight of rats in ND; (B) Body weight of rats in HD. *All exposed groups were compared with control group and only 500 mg/(kg·d) group was significantly different from the control (P < 0.05).
Table Supplementary Table S1. TG Levels (mmol/L) in Adipose tissue of ND Rats
Groups Male Female P Control 1.02 ± 0.08 1.03 ± 0.05 0.97 5 mg/(kg·d) 1.03 ± 0.10 1.03 ± 0.08 0.99 50 mg/(kg·d) 1.21 ± 0.12 1.19 ± 0.20 0.90 500 mg/(kg·d) 1.42 ± 0.16 1.40 ± 0.20 0.95 Table Supplementary Table S2. TG Levels (mmol/L) in Adipose Tissue of HD Rats
Groups Male Female P Control 1.11 ± 0.09 1.13 ± 0.05 0.94 5 mg/(kg·d) 1.09 ± 0.11 1.10 ± 0.15 0.98 50 mg/(kg·d) 1.11 ± 0.20 1.13 ± 0.17 0.94 500 mg/(kg·d) 1.43 ± 0.20 1.40 ± 0.18 0.89 TC and TG levels were detected in liver and adipose (Figure 2A to 2D). Obvious changes in TC or TG levels were not observed, while the TC levels in HD rat livers from all DEHP dose groups were decreased compared to the control. Serum levels of TG, TC, LDL, HDL, LEP, and ADP were also detected (Figure 2E to 2J). In ND, the levels of TC and HDL in 500 mg/(kg·d) group were increased significantly, while there were no evident changes in TG, LDL, LEP, or ADP levels. In HD, the levels of TC, LDL, and HDL in 500 mg/(kg·d) group were significantly higher. LEP levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were increased, whereas levels were decreased in 5 mg/(kg·d) group. ADP levels in all DEHP-treated groups were decreased as compared to the control (P < 0.05).
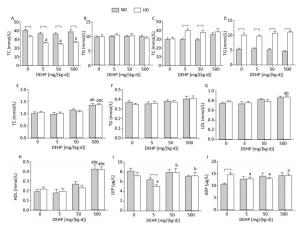
Figure 2. Effects of DEHP and HD on lipids and hormones of rats. (A) TC in liver; (B) TG in liver; (C) TC in adipose; (D) TG in adipose; (E) TC in serum; (F) TG in serum; (G) LDL in serum; (H) HDL in serum; (I) LEP in serum; (J) ADP in serum. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

-
In ND, Stat1, Pparγ, and Aox mRNA levels were significantly increased in 50 mg/(kg·d) and 500 mg/(kg·d) groups. As well, expression of Tyk2 mRNA in 500 mg/(kg·d) group and Fas mRNA in all DEHP-treated groups were significantly elevated. However, Lpl levels did not change significantly among the four dose groups. In HD, Stat1 and Fas levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were also notably higher. Tyk2 and Pparγ mRNA levels in 500 mg/(kg·d) group were higher than the other dose groups, while the Tyk2 levels in 5 mg/(kg·d) and 50 mg/(kg·d) groups were decreased compared with control. In addition, Fas levels in all DEHP–treated groups were significantly increased, while there were no obvious alterations of Lpl expression (P < 0.05) (Figure 3).

Figure 3. Effects of DEHP and HD on expression of related genes in liver. (A) Relative mRNA levels of Tyk2; (B) Relative mRNA levels of Stat1; (C) Relative mRNA levels of Pparγ; (D) Relative mRNA levels of Aox; (E) Relative mRNA levels of Fas; (F) Relative mRNA levels of Lpl; (G) Heat map of relative mRNA levels in liver. The color scale represented the relative mRNA levels, with green indicating down-regulated genes, red indicating up-regulated genes, and the darker the color, the deeper the degree.aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

-
In ND, mRNA levels of Fas in all DEHP-treated groups and Aox in 500 mg/(kg·d) group were significantly higher than in the control group. LC3 mRNA levels in all DEHP-treated groups were increased compared with the control, and the levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were higher than in 5 mg/(kg·d) group. Expression of Tyk2 mRNA in 500 mg/(kg·d) group and Stat1 in all DEHP exposed groups was decreased compared with the control, while Pparγ or Lpl levels were unchanged. In HD, Aox expression in 5 mg/(kg·d) and 500 mg/(kg·d) groups was significantly higher than the control. In contrast, Tyk2 and Pparγ levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were decreased in comparison with control, and expression of Stat1 and Fas in all DEHP-treated groups was lower than in the control. There were no notable changes in Lpl levels among four dose groups (P < 0.05) (Figure 4).
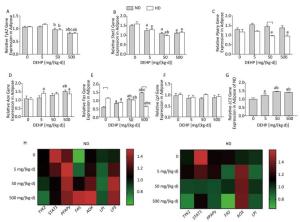
Figure 4. Effects of DEHP and HD on expression of related genes in adipose. (A) Relative mRNA levels of Tyk2; (B) Relative mRNA levels of Stat1; (C) Relative mRNA levels of Pparγ; (D) Relative mRNA levels of Aox; (E) Relative mRNA levels of Fas; (F) Relative mRNA levels of Lpl; (G) Relative mRNA levels of LC3; (H) Heat map of relative mRNA levels in adiposse. The color scale represented the relative mRNA levels, with green indicating down-regulated genes, red indicating up-regulated genes, and the darker the color, the deeper the degree.aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

-
In both diets, HE staining showed that no abnormal changes occurred in liver parenchyma cells of either control group or normal livers. All DEHP-treated groups, especially the 500 mg/(kg·d) group, exhibited structural abnormalities in liver tissue and disordered hepatocyte cords. Vacuolar degeneration and accumulation of inflammatory cytokines was also observed in DEHP-treated groups (Figure 5A).
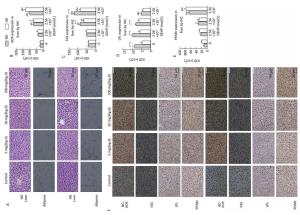
Figure 5. HE staining and Immunohistochemical determination of liver and adipose in ND and HD (400×). (A) HE staining of liver and adipose on ND and HD; (B) IHC staining of AOX expression in liver (400×); (C) IHC staining of FAS expression in liver (400×); (D) IHC staining of LPL expression in liver (400×); (E) IHC staining of PPARγ expression in liver (400×); (F) Immunohistochemistry staining of liver. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

The results showed that the IHC staining intensity of each group of proteins was different. In ND, levels of all proteins of interest in 500 mg/(kg·d) group were significantly increased, with the exception of LPL, which was lower than in other groups. In HD, the expression of AOX, FAS, and PPARr in 500 mg/(kg·d) group was notably higher, while LPL levels among all dose groups were not noticeably changed (P < 0.05) (Figure 5B-F).
Significant alterations in PPARγ levels were not detected in ND. The total TYK2 levels in all DEHP-treated groups were increased compared with control, while the P-TYK2 levels in 5 mg/(kg·d) and 500 mg/(kg·d) groups were decreased. Moreover, the total STAT1 in 500 mg/(kg·d) group was significantly higher than in other groups. However, the P-STAT1 in 50 mg/(kg·d) group was the highest and in 500 mg/(kg·d) group was the lowest. Furthermore, levels of FAS in 50 mg/(kg·d) and 500 mg/(kg·d) groups and AOX in 500 mg/(kg·d) group was all considerably increased. LPL levels in all DEHP-exposed groups were decreased in comparison with control. In HD, the total TYK2 levels in 5 mg/(kg·d) and 500 mg/(kg·d) groups were lower than control, while the P-TYK2 level in 5 mg/(kg·d) group was elevated. In contrast, the total STAT1 in 50 mg/(kg·d) and 500 mg/(kg·d) groups was increased, while the P-STAT1 results were completely opposite. Additionally, PPARγ and FAS protein levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were increased significantly, whereas AOX and LPL levels in all DEHP-treated groups were notably elevated (P < 0.05) (Figure 6).
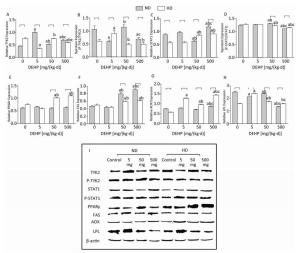
Figure 6. Western blot determination of proteins in liver. (A) The densitometric scans of TYK2/β-actin; (B) The densitometric scans of P-TYK2/TYK2; (C) The densitometric scans of STAT1/β-actin; (D) The densitometric scans of P-STAT1/STAT1; (E) The densitometric scans of PPARγ/β-actin; (F) The densitometric scans of FAS/β-actin; (G) The densitometric scans of AOX/β-actin; (H) The densitometric scans of LPL/β-actin; (I) Western blot assay of liver. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

-
In ND, the total TYK2 in 50 mg/(kg·d) group was increased compared with the control. The P-TYK2 level in 500 mg/(kg·d) group was higher than that of other groups. In comparison with control, the total STAT1 in 500 mg/(kg·d) group was increased and the P-STAT1 levels in 5 mg/(kg·d) and 500 mg/(kg·d) groups were also increased. In addition, PPARγ was higher in all DEHP-exposed groups than the control and the FAS level in 500 mg/(kg·d) group was significantly elevated compared with other groups. LPL expression was highest in the 5 mg/(kg·d) group. There were no obvious changes in AOX levels. The level of autophagy was expressed by LC3II/LC3I, and results demonstrated that the level of autophagy in 50 mg/(kg·d) and 500 mg/(kg·d) groups was significantly higher than that in the control and 5 mg/(kg·d) groups. In HD, the total TYK2 decreased gradually in DEHP-treated groups in comparison with the control. The P-TYK2 level in 50 mg/(kg·d) group was decreased when compared with control. Moreover, the total STAT1 levels in 50 mg/(kg·d) and 500 mg/(kg·d) groups were lower than the control, while the P-STAT1 levels in all DEHP-exposed groups were increased. In comparison with control, the PPARγ and LPL levels in 5 mg/(kg·d) and 50 mg/(kg·d) groups were decreased. Furthermore, FAS in 5 mg/(kg·d) and 500 mg/(kg·d) groups was decreased compared with the control. However, significant alterations of AOX levels were not detected (P < 0.05) (Figure 7).
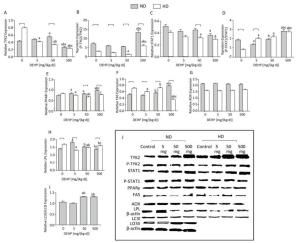
Figure 7. Western blot determination of proteins in adipose. (A) The densitometric scans of TYK2/β-actin; (B) The densitometric scans of P-TYK2/TYK2; (C) The densitometric scans of STAT1/β-actin; (D) The densitometric scans of P-STAT1/STAT1; (E) The densitometric scans of PPARγ/β-actin; (F) The densitometric scans of FAS/β-actin; (G) The densitometric scans of AOX/β-actin; (H) The densitometric scans of LPL/β-actin; (I) The densitometric scans of LC3II/LC3I/β-actin; (J) Western blot assay of adipose. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

-
Factorial analysis demonstrated that HD and DEHP treatment had reciprocal action on TC levels in 5 mg/(kg·d) group, as well as on Fas mRNA levels in 500 mg/(kg·d) group in adipose tissues, and on mRNA levels of Pparγ in 50 mg/(kg·d) group, Aox in 5 mg/(kg·d) group, and Tyk2 in 500 mg/(kg·d) group in liver tissues. Thus, the above results indicate the joint effects of HD and DEHP (Table 2).
Table 2. The Joint Effects of HD and DEHP on the Indexes of Lipid Metabolism
Indexes HD DEHP HD + DEHP F P F P F P TC of adipose in 5 mg/(kg·d) group 7.75 0.007 6.11 0.017 5.93 0.018 Fas mRNA of adipose in 500 mg/(kg·d) group 20.14 < 0.001 5.61 0.025 15.99 < 0.001 Pparγ mRNA of liver in 50 mg/(kg·d) group 20.76 < 0.001 9.40 0.005 8.57 0.006 Aox mRNA of liver in 5 mg/(kg·d) group 164.57 < 0.001 56.27 < 0.001 76.34 < 0.001 Tyk2 mRNA of liver in 500 mg/(kg·d) group 11.78 0.002 15.87 < 0.001 12.69 0.001 AOX of adipose in 500 mg/(kg·d) group by IHC 5.91 0.041 88.99 < 0.001 21.32 0.002 FAS of adipose in 50 mg/(kg·d) group by IHC 35.17 < 0.001 72.32 < 0.001 8.93 0.017 FAS of adipose in 500 mg/(kg·d) group by IHC 36.11 < 0.001 177.67 < 0.001 7.55 0.025 LPL of adipose in 5 mg/(kg·d) group by IHC 23.71 0.001 23.97 0.001 42.12 < 0.001 LPL of adipose in 50 mg/(kg·d) group by IHC 46.30 < 0.001 58.70 < 0.001 42.43 < 0.001 -
As an organic environmental pollutant, DEHP has adverse effects on the reproductive system, carcinogenesis, and lipid metabolism[17]. As well, many studies suggest that JAK/STAT may regulate leptin-induced abnormal lipid metabolism[18]. As well, autophagy is currently known to regulate lipid balance in adipose tissue by regulating adipocyte differentiation[19]. A previous study in our laboratory found that DEHP can influence the expression of lipid metabolism related markers by regulating JAK3 and STAT5A in adolescent rats[20]. However, Yiyang et al., only observed lipid metabolism levels for 4 weeks of DEHP exposure, which did not cover the entire puberty of the rats. As puberty is a critical period of fat development in rats, we extended the exposure time to 8 weeks, which encompasses the entire pubescent period in rats. Furthermore, previous studies have shown that a high-fat diet may have joint toxicity with phthalates[9, 10].
Recent studies have shown that TYK2 impacts the regulation of energy balance. Our study demonstrated decreased TYK2 levels in adipose. TYK2 promotes differentiation and maturation of brown fat. Derecka et al.[21] found that TYK2 knockout mice displayed many metabolic abnormalities, including obesity. Significant elevation of P-TYK2 in adipose of the high-dose group suggests that it may be the total TYK2, and not P-TYK2, which plays a vital role in lipid metabolism. In contrast, increased TYK2 in liver suggested that it promoted the depletion of energy in liver, whereas the reduced P-TYK2 suggests possible regulation of decompensation, as evidenced by the high expression of adipogenic enzymes.
Previous studies suggest that STAT1 functions in lipogenesis and lipid uptake[22]. Our data showed elevated P-STAT1 levels in adipose of DEHP-treated groups from both diets, suggesting that DEHP exposure activated STAT1, causing the formation and accumulation of lipids in adipose tissue. Decrease of P-STAT1 in the liver may be the result of compensatory regulation. However, it is also possible that the function of STAT1 is tissue-specific. Lee et al.[23] indicated that IFN-α, which was a key immunoregulatory cytokine, inhibited the accumulation of lipid droplets and the expression of adipogenesis related genes in the early stages of fat cell development. Our results regarding P-STAT1 levels in the liver may suggest that DEHP exposure prevented P-STAT1 from inhibiting expression of PPARγ and C/EBPα, which are the key cytokines related to adipogenesis in liver.
When exposed to foreign compounds, such as DEHP, the role of autophagy is to attempt to maintain body stability through energy redistribution, protein remodeling, and hormone regulation. Our results showed that LC3 gene and protein levels in adipose of DEHP-treated groups were increased compared with the control. Many studies have confirmed that the activity of autophagy in adipose is significantly elevated under obese conditions, and autophagy markers are significantly associated with obesity[24]. Kosacka et al., found that the level of LC3, an autophagy protein, was significantly increased in adipose of type 2 diabetes patients and obese people, and the expression of Atg5/12 autophagy complex protein was 2 times higher than that of the control group. Expression of mRNA and corresponding proteins of autophagy related genes also display a difference in adipose from obese patients and healthy adipose[8]. The current study is an initial exploration of whether autophagy plays a role in lipid metabolism. Our results suggest that autophagy is involved in DEHP-induced lipid metabolism disorders in rats. The specific mechanism by which autophagy affects lipid metabolism will be explored in depth in our future research.
Expression of PPARγ was not in accordance with activated STAT1 in our results, suggesting that the PPARγ expression in the liver may not occur downstream of the STATs. Studies have found that the increases in PPARγ expression occurred before any effects in body weight and fat mass, suggesting that transcription factors such as STATs may be responsible for the accelerated accumulation of adipose tissue. As well, PPARγ may also play a role in inhibition of the JAK/STAT pathway in the leptin signaling pathway, thereby further blocking the inhibitory effect of leptin on insulin secretion[25].
Previous research[26] has referred to the inhibition of LPL and PPARγ2 by IFNγ-activated STAT1 in Murine models. Our results suggest that LPL levels gradually decreased in DEHP-treated, ND, livers, which was consistent with P-STAT1 levels, indicating that the liver was actively regulated to reduce the uptake and storage of TG. In adipose, DEHP exposure can elevate AOX expression. Previous studies have confirmed that liver has the most abundant Aox mRNA and protein expression., and that Aox-/- mice have steatohepatitis and spontaneous peroxisome proliferation[27]. In contrast, elevated STAT5A levels may occur in the liver after DEHP exposure, causing increased AOX levels and further affecting hepatic lipid metabolism and pathogenesis of liver fat[28]. The significantly increased FAS levels suggest that fatty acid synthesis was elevated in liver.
In the present study, the body weight of rats in 500 mg/(kg·d) group was the highest in both diets. However, Chen et al., did not find obvious changes in rat body weight after DEHP exposure[29], we suggest that this discrepancy may be due to the age of the rats. Chen et al., exposed rats weighing 110-160 g to DEHP. Although rats of this age are not at a critical stage of fat development, body weight tended to be stable. In addition, another study which exposed rats weighing 24-26 g to DEHP for 5 weeks revealed that the weight in 50 and 200 mg/(kg·d) groups initially increased, and then decreased[30]. Our study revealed many different trends in lipid metabolism indicators between ND and HD rats. In the case of HD, these trends may due to increased sensitivity or insensitivity to certain indicators of changes in rats. Alternatively, it may be that the endocrine process was completely disrupted in rats subjected to HD treatment. HD intake in DEHP-exposed male rats increased energy intake and body weight and led to glucose tolerance[10]. On this basis, we determined that DEHP may have a joint effect with HD on TC levels in adipose, based on mRNA expression of Fas in adipose, Pparγ, Aox and Tyk2 mRNA expression in liver, as well as immunohistochemistry derived protein expression of AOX, FAS, and LPL in adipose. This may prove that a HD, in conjunction with DEHP, plays a role in lipid metabolism, leading to lipid metabolism disorders and obesity. However, the trend of changes in doses did not agree. To further investigate the effect of DEHP combined with high-fat diet on lipid metabolism, the study will be expanded to investigate the roles of additional pathway molecules.
DEHP exposure resulted in changes in the expression of TYK2/STAT1 in rats, affected the level of autophagy, and further affected the expression of lipid metabolism-related factors regulated by TYK2/STAT1 and autophagy, leading to disorder of lipid metabolism and ultimately obesity. In conclusion, exposure to environmental DEHP causes dyslipidemia, abnormal weight gain, and subsequent obesity in rats. This abnormality may due to activation of TYK2/STAT1 which leads to further alteration of enzyme levels and results in increased adipogenesis, lipid accumulation, and decreased lipid hydrolysis. Additionally, autophagy may play a potential role during this process. High-fat diet has an important impact on lipid metabolism. Our study suggests that high-fat diet may have joint effects with DEHP in lipid metabolism. The specific mechanism underlying this joint effect requires further study. In the future, we will further verify the role of TYK2/STAT1 and autophagy in lipid metabolism disorders caused by DEHP in vitro.
-
The authors declare that they have no conflict of interest.
doi: 10.3967/bes2019.055
Di (2-ethylhexyl) phthalate Disorders Lipid Metabolism via TYK2/STAT1 and Autophagy in Rats
-
Abstract:
Objective Previous studies have indicated that the plasticizer di (2-ethylhexyl) phthalate (DEHP) affects lipid accumulation; however, its underlying mechanism remains unclear. We aim to clarify the effect of DEHP on lipid metabolism and the role of TYK2/STAT1 and autophagy. Methods In total, 160 Wistar rats were exposed to DEHP[0, 5, 50, 500 mg/(kg·d)] for 8 weeks. Lipid levels, as well as mRNA and protein levels of TYK2, STAT1, PPARγ, AOX, FAS, LPL, and LC3 were detected. Results The results indicate that DEHP exposure may lead to increased weight gain and altered serum lipids. We observed that DEHP exposure affected liver parenchyma and increased the volume or number of fat cells. In adipose tissue, decreased TYK2 and STAT1 promoted the expression of PPARγ and FAS. The mRNA and protein expression of LC3 in 50 and 500 mg/(kg·d) groups was increased significantly. In the liver, TYK2 and STAT1 increased compensatorily; however, the expression of FAS and AOX increased, while LPL expression decreased. Joint exposure to both a high-fat diet and DEHP led to complete disorder of lipid metabolism. Conclusion It is suggested that DEHP induces lipid metabolism disorder by regulating TYK2/STAT1. Autophagy may play a potential role in this process as well. High-fat diet, in combination with DEHP exposure, may jointly have an effect on lipid metabolism disorder. -
Key words:
- DEHP /
- TYK2 /
- STAT1 /
- High-fat diet /
- Autophagy /
- Lipid metabolism
-
Figure 2. Effects of DEHP and HD on lipids and hormones of rats. (A) TC in liver; (B) TG in liver; (C) TC in adipose; (D) TG in adipose; (E) TC in serum; (F) TG in serum; (G) LDL in serum; (H) HDL in serum; (I) LEP in serum; (J) ADP in serum. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Figure 3. Effects of DEHP and HD on expression of related genes in liver. (A) Relative mRNA levels of Tyk2; (B) Relative mRNA levels of Stat1; (C) Relative mRNA levels of Pparγ; (D) Relative mRNA levels of Aox; (E) Relative mRNA levels of Fas; (F) Relative mRNA levels of Lpl; (G) Heat map of relative mRNA levels in liver. The color scale represented the relative mRNA levels, with green indicating down-regulated genes, red indicating up-regulated genes, and the darker the color, the deeper the degree.aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Figure 4. Effects of DEHP and HD on expression of related genes in adipose. (A) Relative mRNA levels of Tyk2; (B) Relative mRNA levels of Stat1; (C) Relative mRNA levels of Pparγ; (D) Relative mRNA levels of Aox; (E) Relative mRNA levels of Fas; (F) Relative mRNA levels of Lpl; (G) Relative mRNA levels of LC3; (H) Heat map of relative mRNA levels in adiposse. The color scale represented the relative mRNA levels, with green indicating down-regulated genes, red indicating up-regulated genes, and the darker the color, the deeper the degree.aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Figure 5. HE staining and Immunohistochemical determination of liver and adipose in ND and HD (400×). (A) HE staining of liver and adipose on ND and HD; (B) IHC staining of AOX expression in liver (400×); (C) IHC staining of FAS expression in liver (400×); (D) IHC staining of LPL expression in liver (400×); (E) IHC staining of PPARγ expression in liver (400×); (F) Immunohistochemistry staining of liver. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Figure 6. Western blot determination of proteins in liver. (A) The densitometric scans of TYK2/β-actin; (B) The densitometric scans of P-TYK2/TYK2; (C) The densitometric scans of STAT1/β-actin; (D) The densitometric scans of P-STAT1/STAT1; (E) The densitometric scans of PPARγ/β-actin; (F) The densitometric scans of FAS/β-actin; (G) The densitometric scans of AOX/β-actin; (H) The densitometric scans of LPL/β-actin; (I) Western blot assay of liver. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Figure 7. Western blot determination of proteins in adipose. (A) The densitometric scans of TYK2/β-actin; (B) The densitometric scans of P-TYK2/TYK2; (C) The densitometric scans of STAT1/β-actin; (D) The densitometric scans of P-STAT1/STAT1; (E) The densitometric scans of PPARγ/β-actin; (F) The densitometric scans of FAS/β-actin; (G) The densitometric scans of AOX/β-actin; (H) The densitometric scans of LPL/β-actin; (I) The densitometric scans of LC3II/LC3I/β-actin; (J) Western blot assay of adipose. aComparison with control (P < 0.05); bComparison with 5 mg/(kg·d) group (P < 0.05); cComparison with 50 mg/(kg·d) group (P < 0.05);

Table 1. The Primers of Real Time Quantitative PCR
Gene Name Primer Sequence Tyk2 Forward 5'-ACTATGCATCTGATGTCTGGTCCTT-3' Reverse 5'-CAGTCATCTGGCCCTGGGTTA-3' Stat1 Forward 5'-TGGCAGCTGAGTTCCGACA-3' Reverse 5'-CTGGCACAACTGGGTTTCAAAG-3' Pparγ Forward 5'-GGAGCCTAAGTTTGAGTTTGCTGTG-3' Reverse 5'-TGCAGCAGGTTGTCTTGGATG-3' Fas Forward 5'-TGGTCACAGACGATGACAGGA-3' Reverse 5'-AGGCGTCGAACTTGGACAGA-3' Aox Forward 5'-ATTGGCACCTACGCCCAGAC-3' Reverse 5'-CCAGGCCACCACTTAATGGAA-3' Lpl Forward 5'-CATTGGAGAAGCCATTCGTGT-3' Reverse 5'-TGCCTTGCTGGGGTTTTC-3' LC3 Forward 5'-CGTCCTGGACAAGACCAAGT-3' Reverse 5'-TCTCGCTCTCGTACACTTCA-3' Supplementary Table S1. TG Levels (mmol/L) in Adipose tissue of ND Rats
Groups Male Female P Control 1.02 ± 0.08 1.03 ± 0.05 0.97 5 mg/(kg·d) 1.03 ± 0.10 1.03 ± 0.08 0.99 50 mg/(kg·d) 1.21 ± 0.12 1.19 ± 0.20 0.90 500 mg/(kg·d) 1.42 ± 0.16 1.40 ± 0.20 0.95 Supplementary Table S2. TG Levels (mmol/L) in Adipose Tissue of HD Rats
Groups Male Female P Control 1.11 ± 0.09 1.13 ± 0.05 0.94 5 mg/(kg·d) 1.09 ± 0.11 1.10 ± 0.15 0.98 50 mg/(kg·d) 1.11 ± 0.20 1.13 ± 0.17 0.94 500 mg/(kg·d) 1.43 ± 0.20 1.40 ± 0.18 0.89 Table 2. The Joint Effects of HD and DEHP on the Indexes of Lipid Metabolism
Indexes HD DEHP HD + DEHP F P F P F P TC of adipose in 5 mg/(kg·d) group 7.75 0.007 6.11 0.017 5.93 0.018 Fas mRNA of adipose in 500 mg/(kg·d) group 20.14 < 0.001 5.61 0.025 15.99 < 0.001 Pparγ mRNA of liver in 50 mg/(kg·d) group 20.76 < 0.001 9.40 0.005 8.57 0.006 Aox mRNA of liver in 5 mg/(kg·d) group 164.57 < 0.001 56.27 < 0.001 76.34 < 0.001 Tyk2 mRNA of liver in 500 mg/(kg·d) group 11.78 0.002 15.87 < 0.001 12.69 0.001 AOX of adipose in 500 mg/(kg·d) group by IHC 5.91 0.041 88.99 < 0.001 21.32 0.002 FAS of adipose in 50 mg/(kg·d) group by IHC 35.17 < 0.001 72.32 < 0.001 8.93 0.017 FAS of adipose in 500 mg/(kg·d) group by IHC 36.11 < 0.001 177.67 < 0.001 7.55 0.025 LPL of adipose in 5 mg/(kg·d) group by IHC 23.71 0.001 23.97 0.001 42.12 < 0.001 LPL of adipose in 50 mg/(kg·d) group by IHC 46.30 < 0.001 58.70 < 0.001 42.43 < 0.001 -
[1] Kimber I, Dearman RJ. An assessment of the ability of phthalates to influence immune and allergic responses. Toxicology, 2010; 271, 73-82. doi: 10.1016/j.tox.2010.03.020 [2] North ML, Takaro TK, Diamond ML, et al. Effects of phthalates on the development and expression of allergic disease and asthma. Ann Allergy Asthma Immunol, 2014; 112, 496-502. doi: 10.1016/j.anai.2014.03.013 [3] Lai FN, Liu JC, Li L, et al. Di (2-ethylhexyl) phthalate impairs steroidogenesis in ovarian follicular cells of prepuberal mice. Arch Toxicol, 2017; 91, 1279-92. doi: 10.1007/s00204-016-1790-z [4] Schmidt JS, Schaedlich K, Fiandanese N, et al. Effects of di(2-ethylhexyl) phthalate (DEHP) on female fertility and adipogenesis in C3H/N mice. Environ Health Perspect, 2012; 120, 1123-9. doi: 10.1289/ehp.1104016 [5] Feige JN, Gelman L, Rossi D, et al. The endocrine disruptor monoethyl-hexyl-phthalate is a selective peroxisome proliferator-activated receptor gamma modulator that promotes adipogenesis. J Biol Chem, 2007; 282, 19152-66. doi: 10.1074/jbc.M702724200 [6] Kloting N, Hesselbarth N, Gericke M, et al. Di-(2-Ethylhexyl)-Phthalate (DEHP) Causes Impaired Adipocyte Function and Alters Serum Metabolites. PLoS one, 2015; 10, e0143190. doi: 10.1371/journal.pone.0143190 [7] Ji J, Petropavlovskaia M, Khatchadourian A, et al. Type 2 diabetes is associated with suppression of autophagy and lipid accumulation in beta-cells. J Cell Mol Med, 2019; 23, 2890-900. doi: 10.1111/jcmm.2019.23.issue-4 [8] Kosacka J, Kern M, Kloting N, et al. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol, 2015; 409, 21-32. doi: 10.1016/j.mce.2015.03.015 [9] Serrano SE, Braun J, Trasande L, et al. Phthalates and diet:a review of the food monitoring and epidemiology data. Environ Health, 2014; 13, 43. doi: 10.1186/1476-069X-13-43 [10] Strakovsky RS, Lezmi S, Shkoda I, et al. In utero growth restriction and catch-up adipogenesis after developmental di (2-ethylhexyl) phthalate exposure cause glucose intolerance in adult male rats following a high-fat dietary challenge. J Nutr Biochem, 2015; 26, 1208-20. doi: 10.1016/j.jnutbio.2015.05.012 [11] Wu K, Tan XY, Xu YH, et al. JAK family members:Molecular cloning, expression profiles and their roles in leptin influencing lipid metabolism in Synechogobius hasta. Comp Biochem Physiol B Biochem Mol Biol, 2017; 203, 122-31. doi: 10.1016/j.cbpb.2016.10.004 [12] Richard AJ, Stephens JM. The role of JAK-STAT signaling in adipose tissue function. Biochimica Biophys Acta, 2014; 1842, 431-9. doi: 10.1016/j.bbadis.2013.05.030 [13] Stephens JM, Morrison RF, Pilch PF. The expression and regulation of STATs during 3T3-L1 adipocyte differentiation. J Biol Chem, 1996; 271, 10441-4. doi: 10.1074/jbc.271.18.10441 [14] Richard AJ, Stephens JM. Emerging roles of JAK-STAT signaling pathways in adipocytes. Trends Endocrinol Metab, 2011; 22, 325-32. doi: 10.1016/j.tem.2011.03.007 [15] Wu X, Jiang L, Sun X, et al. Mono(2-ethylhexyl) phthalate induces autophagy-dependent apoptosis through lysosomal-mitochondrial axis in human endothelial cells. Food Chem Toxicol, 2017; 106, 273-82. doi: 10.1016/j.fct.2017.05.069 [16] Issemann I, Green S. Activation of a member of the steroid hormone receptor superf amily by peroxisome prolif erators. Nature, 1990; 347, 645-50. doi: 10.1038/347645a0 [17] Rowdhwal SSS, Chen J. Toxic Effects of Di-2-ethylhexyl Phthalate:An Overview. Biomed Res Int, 2018; 2018, 1750368. [18] Song YF, Wu K, Tan XY, et al. Effects of recombinant human leptin administration on hepatic lipid metabolism in yellow catfish Pelteobagrus fulvidraco:in vivo and in vitro studies. Gen Comp Endocrinol, 2015; 212, 92-9. doi: 10.1016/j.ygcen.2015.01.022 [19] Singh R, Xiang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest, 2009; 119, 3329-39. [20] Jia Y, Liu T, Zhou L, et al. Effects of Di-(2-ethylhexyl) Phthalate on Lipid Metabolism by the JAK/STAT Pathway in Rats. Int J Environ Res Public Health, 2016; 13, 1085. doi: 10.3390/ijerph13111085 [21] Derecka M, Gornicka A, Koralov SB, et al. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab, 2012; 16, 814-24. doi: 10.1016/j.cmet.2012.11.005 [22] Hao J, Zhang YJ, Lv X, et al. IFN-gamma induces lipogenesis in mouse mesangial cells via the JAK2/STAT1 pathway. Am J Physiol Cell Physiol, 2013; 304, C760-7. doi: 10.1152/ajpcell.00352.2012 [23] Lee K, Um SH, Rhee DK, et al. Interferon-alpha inhibits adipogenesis via regulation of JAK/STAT1 signaling. Biochimica Biophys Acta, 2016; 1860, 2416-27. doi: 10.1016/j.bbagen.2016.07.009 [24] Kovsan J, Bluher M, Tarnovscki T, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab, 2011; 96, E268-77. doi: 10.1210/jc.2010-1681 [25] Mynatt RL, Stephens JM. Agouti regulates adipocyte transcription factors. Am J Physiol Cell Physiol, 2001; 280, C954-61. doi: 10.1152/ajpcell.2001.280.4.C954 [26] Zhao P, Stephens JM. Identification of STAT target genes in adipocytes. Jak-stat, 2013; 2, e23092. doi: 10.4161/jkst.23092 [27] Fan CY, Pan J, Usuda N, et al. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem, 1998; 273, 15639-45. doi: 10.1074/jbc.273.25.15639 [28] Coulter AA, Stephens JM. STAT5 activators modulate acyl CoA oxidase (AOX) expression in adipocytes and STAT5A binds to the AOX promoter in vitro. J Biol Chem, 2006; 344, 1342-5. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=5bdc24aec34f356475aa59057e46e467 [29] Chen H, Zhang W, Rui BB, et al. Di(2-ethylhexyl) phthalate exacerbates non-alcoholic fatty liver in rats and its potential mechanisms. Environ Toxicol Pharmacol, 2016; 42, 38-44. doi: 10.1016/j.etap.2015.12.016 [30] Lv Z, Cheng J, Huang S, et al. DEHP induces obesity and hypothyroidism through both central and peripheral pathways in C3H/He mice. Obesity, 2016; 24, 368-78. doi: 10.1002/oby.21359 -




 下载:
下载:




 Quick Links
Quick Links