-
Brucella is gram-negative, facultative, intracellular bacteria implicated in infectious zoonosis diseases, particularly among domestic animals, that are also transmittable to humans. The genus Brucella is classified based on the primary host preferences, pathogenicity, host preference, and phenotypic characteristics of its species[1]. The clinical features of brucellosis overlap with those of an extensive range of infectious and non-infectious diseases; therefore, laboratory testing is deemed the most reliable approach to diagnose this infection[2]. Microbiological culturing and serological examinations are the most common methods for the detection of Brucella. Although the isolation of these bacteria is the ‘gold standard’ approach, the microbial culture often gives false negative results and dependent on the culture medium, the quantity of the circulating bacteria, and the species of Brucella[3]. Therefore, serological tests such as the standard serum agglutination test (SAT) seem to be more effective for diagnosis, although an occasional case of cross-reaction or false-positive reaction in the samples from areas with subclinical prevalence of brucellosis has been promoted[4]. The present study is a comprehensive analyses of the genomic nucleotide sequences of Brucella spp
. that also developed a nested-polymerase chain reaction (PCR) assay for the identification and genotyping of Brucella, especially for those contained in blood specimens. A total of 36 reference strains (B. abortus biovars[1-7], B. melitensis biovars[1-3], B. suis biovars[1-5], B. canis; B. ovis; B. neotomae, B. pinnipedialis, and B. microti) were used. All strains of B. abortus, B. melitensis, B. suis, B. canis; B. ovis; and B. neotomae were preserved in Brucellosis Laboratory, CDC, China. The information on the DNA of B. ceti, B. microti, and B. pinnipedialis strains were sourced from the US National Center for Biotechnology Information (NCBI) website. The Rev1, M5, and M28 belonged to the B. melitensis vaccine strains; 104M and S19 to the B. abortus vaccine strains; VBI22 and VacciS2 to the B. suis vaccine strains; and 45/20 and B1119 to the B. abortus rough strains.
A total of 89 clinical strains isolated from the patient's blood samples were typed through the conventional biological methods. A total of 7 blood and sera samples were collected from the patients and sheep. The B114, B115, and lanzhou-26 were clinical human anticoagulant whole blood samples. The serum antibody titer was > 1:100. However, no Brucella species was isolated. The B243, B251, and B252 samples were collected from the sera of sheep from a farm. The B. melitensis biovars 3 strains was isolated from B252 sample. B. canis strain was isolated from BJ10 samples collected from anticoagulant whole blood samples from dogs at the farm. The samples were positive for Brucella antibodies (Table 1)[5].
Table 1. The 36 reference strains, 89 clinical isolates, and 7 blood and sera samples used in this study
NO. Strain ID Biovars Note NO. Strain D Biovars Note NO. Strain D Biovars Note NO. Strain D Biovars Note 1 A13334 A1 REF 34 63/290 Ovis REF 67 13106 M3 Isolate 100 78037 M2 Isolate 2 2308 A1 REF 35 25840 Ovis REF 68 13107 M3 Isolate 101 79029 M3 Isolate 3 9-941 A1 REF 36 B2-94 Pinni REF 69 13108 M3 Isolate 102 79054 M3 Isolate 4 86/8/59 A2 REF 37 208 S1 Isolate 70 13138 S1 Isolate 103 79077 M3 Isolate 5 Tulya A3 REF 38 226 S3 Isolate 71 13139 S2 Isolate 104 79104 M3 Isolate 6 292 A4 REF 39 1048 A3 Isolate 72 13147 S1 Isolate 105 79130 M3 Isolate 7 B3196 A5 REF 40 1057 A1 Isolate 73 13160 A1 Isolate 106 80345 M3 Isolate 8 870 A6 REF 41 1148 A3 Isolate 74 13195 M3 Isolate 107 80347 M3 Isolate 9 63/75 A7 REF 42 3001 M3 Isolate 75 13248 M2 Isolate 108 80355 M2 Isolate 10 C86 A9 REF 43 5017 M3 Isolate 76 13331 S2 Isolate 109 80356 M2 Isolate 11 104M A Vacci 44 7033 M3 Isolate 77 13332 S1 Isolate 110 80359 A3 Isolate 12 S19 A Vacci 45 8257 A1 Isolate 78 13341 M3 Isolate 111 80392 A9 Isolate 13 45/20 A REF 46 8416 A9 Isolate 79 23385 Canis Isolate 112 81006 M3 Isolate 14 B1119 A REF 47 9057 S3 Isolate 80 52141 Canis Isolate 113 81011 A7 Isolate 15 16M M1 REF 48 10009 S3 Isolate 81 57002 A3 Isolate 114 81033 M3 Isolate 16 63/9 M2 REF 49 10036 Canis Isolate 82 60033 A1 Isolate 115 84001 M3 Isolate 17 Eher M3 REF 50 10062 S3 Isolate 83 61005 M3 Isolate 116 86028 M1 Isolate 18 NI M REF 51 11038 Canis Isolate 84 63006 M2 Isolate 117 86028 S1 Isolate 19 M5 M Vacci 52 11055 A3 Isolate 85 63031 A1 Isolate 118 88040 S1 Isolate 20 M28 M Vacci 53 11059 M3 Isolate 86 64008 S3 Isolate 119 90001 A3 Isolate 21 Rev1 M Vacci 54 12088 M3 Isolate 87 64027 M2 Isolate 120 92008 M3 Isolate 22 1330 S1 REF 55 12099 M3 Isolate 88 64029 A1 Isolate 121 B257 A1 Isolate 23 Thomsen S2 REF 56 12110 M3 Isolate 89 65050 M2 Isolate 122 HN-13 M1 Isolate 24 686 S3 REF 57 13031 M3 Isolate 90 65066 M2 Isolate 123 HNLG M3 Isolate 25 40 S4 REF 58 13034 M3 Isolate 91 65079 A1 Isolate 124 HNLR M3 Isolate 26 513 S5 REF 59 13051 M3 Isolate 92 66148 M2 Isolate 125 HNMB M3 Isolate 27 VacciS2 S Vacci 60 13067 M3 Isolate 93 72003 M2 Isolate 126 B114 / Blood 28 VBI22 S Vacci 61 13080 M3 Isolate 94 73011 M3 Isolate 127 B115 A1 Blood 29 RM6/66 Canis REF 62 13081 M3 Isolate 95 73096 S3 Isolate 128 BJ10 Canis Blood 30 10759 Ceti REF 63 13083 M3 Isolate 96 73099 M3 Isolate 129 Lanzhou-26 A1 Blood 31 28753 Ceti REF 64 13096 S3 Isolate 97 73101 M3 Isolate 130 B243 S3 Serum 32 4915 Micro REF 65 13102 M3 Isolate 98 73162 M3 Isolate 131 B251 / Serum 33 5K33 Neoto REF 66 13105 M3 Isolate 99 78014 M3 Isolate 132 B252 M3 Serum Note. A: B. abortus biovar, M: B. melitensis biovar, S: B. suis biovar, Canis: B. canis, Ceti: B. ceti, Pinni: B. pinnipedialis, Neoto: B. neotomae, Ovis: B. ovis, Micro: B. microti, REF: Reference strain, Isolate: Clinical isolate. /: Brucella was not detected in the blood samples by culturing or nested-PCR assay. REF, reference. A total of 7 non-Brucella DNA were used to verify the specificity of the nested-PCR assay, which included Bacillus cereus, Bacillus anthracis, Escherichia coli o:157, Salmonella, Pseudomonas aeruginosa, Yersinia enterocolitica o:9, and Vibrio cholerae. E. coli o:157 strain was treated as a negative quality control in this study.
The Bacterial Genomic DNA Extraction Kit (Spin column; Tiangen Biotechnology [Beijing] Co., Ltd, China) was used to extract the nucleic acid DNA from the reference strains and the clinical isolates. Blood and serum samples were extracted by using this kit.
Comparative analysis of the whole genome sequences of Brucella spp. from the NCBI databases revealed that the nucleotide sequences in transposase IS711 orfA and orfB of Brucella spp.
possess nucleotide polymorphisms and contain Brucella species and biovars specificity. The oligonucleotide primers were designed based on orfA and orfB (Figure 1). A nested-PCR assay was established to identify and genotype the strains of Brucella spp. The primers used included F1: 5′-AACGTAACCATACATAGCGCATG-3′ and R1: 5′-ACAGATGAGCAATGGAACCGGAT-3′ and F2: 5′-GAATGGGTGCAATTTCTCGC-3′ and R2: 5′-ATATCTTCCGGGGCGAGTTGGTA-3′. The first PCR was conducted with the primer pairs F1 and R1 using the extracted DNA as the template. The second PCR was conducted with the primer pairs F2 and R2 using the first PCR amplicons as the template. 
Figure 1. The nucleotide polymorphisms in transposase IS711 orfA and orfB of Brucella spp.* and vertical bar with a blue background: the base mutation site.
The first PCR reaction system included 2×Taq mastermix (12.5 μL, primer F1, and primer R1 [10 μmol/L]; Kangwei Century Biotechnology Co., Ltd, China) 0.6 μL each and DNA template 2 μL, to which 9.3–25.0 μL of distilled water was added. The first PCR reaction conditions were as follows: 94 ℃ for 4 min; 94 ℃ for 45 s, 55 ℃ for 45 s, and 72 ℃ for 60 s, 30 cycles. The final extension was performed at 72 ℃ for 5 min. The second PCR reaction system and reaction conditions were the same as for the first PCR system, except for the template DNA from the first PCR amplicons (nested-PCR). A bright electrophoresis band of approximately 666 bp was amplified for the strains of Brucella spp. by using the nested-PCR assay. A total of 7 non-Brucella DNA could not be amplified by nested-PCR assay. The nested-PCR amplicons were purified and sequenced. Multiple sequences alignments were performed using the Clustal W, and a schematic representation of each locus was generated using the MEGA 5.1 by using the unweighted group average method (UPGMA), Neighbor-Joining tree construction, and Tamura-Nei algorithm with 1,000 bootstraps.
The sensitivity of the nested-PCR assay was determined by using 36 reference strains. All strains could amplify the positive bands. The minimum detection limit was tested by using 2-fold decreasing dilutions of B. suis 1330 DNA. The initial DNA (56.2 ng/μL) was diluted to 0.03 fg/μL. After several testing, the sensitivity of the nested-PCR was 3.35 fg, which was equivalent to 1 copy number of Brucella DNA in a 25-μL reaction system, considering that 24-fg nucleic acid DNA equals to approximately 7 copies of Brucella DNA[6].
A total of 36 Brucella reference strains, 89 clinical isolates, and 7 blood samples were examined by nested-PCR assay. The nested-PCR products were sequenced, and the sequences were clustered (Figure 2). The cluster analysis was performed as follows:
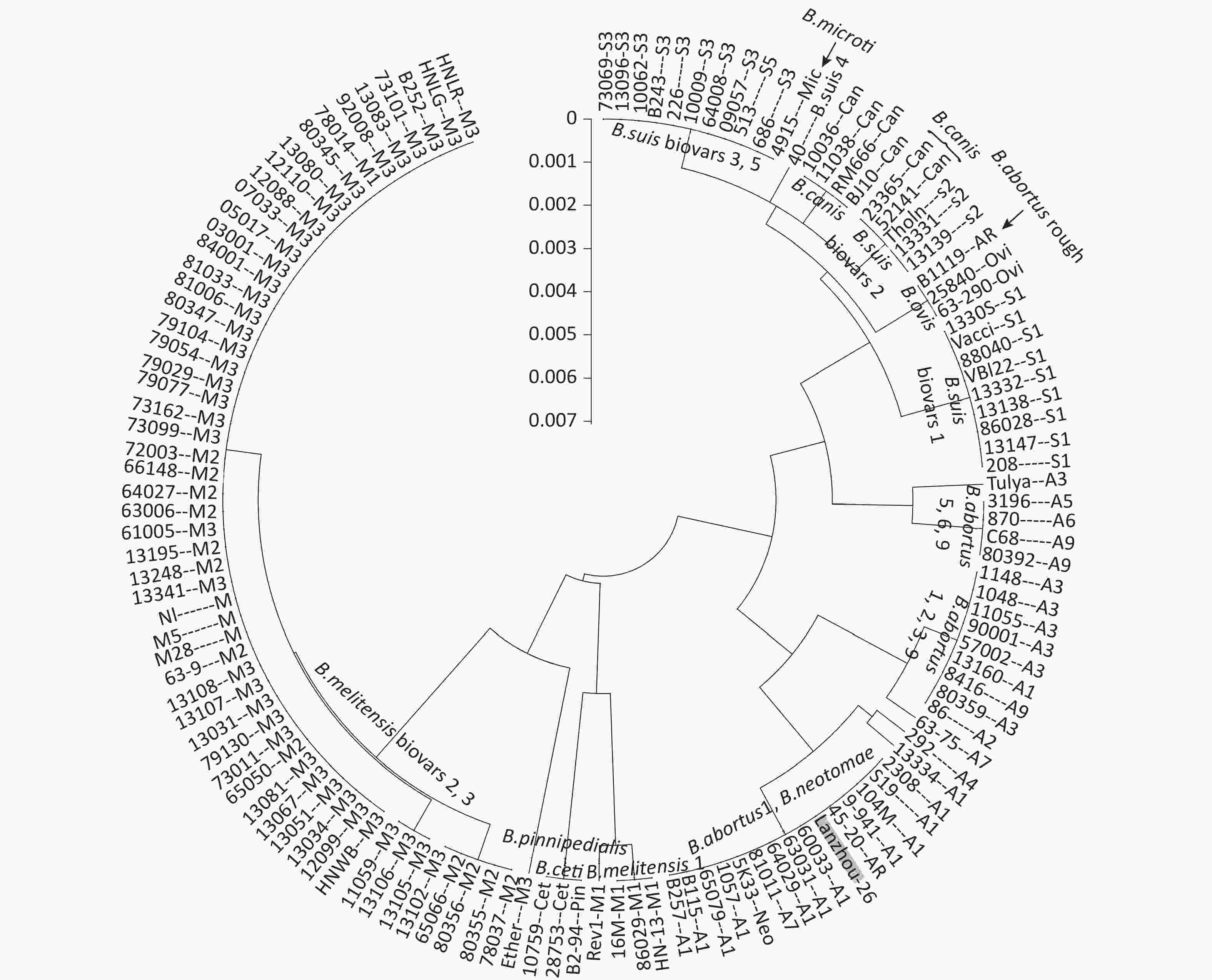
Figure 2. The cluster analysis of 36 Brucella reference strains, 89 clinical isolates, and 7 blood samples by nested-PCR. The black background of the code numbers indicates blood samples (B115, B243, B252, BJ10, and Lanzhou-26) in Figure 1.
1) Three strains isolated from marine animals were assigned to 1 group, which included B. ceti 10759, B. ceti 28753, and B. pinnipedialis B2–94.
2) All B. melitensis biovar 2 and B. melitensis biovar 3 strains were clustered together.
3) All B. abortus biovar 1 strains were clustered together, along with 1 B. neotomae 5K33 strain.
4) The B. ovis 63/290 and B. ovis 25840 strains were clustered together, along with 1 B. abortus B1119 rough strains.
5) All B. suis biovar 3 strains were clustered together, along with 1 B. microti 4915 strain.
6) A total of 6 B. canis strains were clustered into 2 groups, 1 group included 4 B. canis strains, while another included 2 B. canis strains along with 3 B. suis biovar 2 strains.
7) The 7 blood samples could not be amplified with a single primer pairs (F1 & R1 or F2 & R2). However, 5 blood samples showed bright electrophoresis bands through the nested-PCR assay with the primer pairs (F1 & R1 → F2 & R2). The cluster analysis revealed that B115 was B. abortus biovar 1, B252 was B. melitensis biovar 3, B243 was B. suis biovar 3, BJ10 was B. canis, and Lanzhou-26 was B. abortus biovar 1 (Figure 3). All 7 blood samples could not be amplified with a single primer B4 and B5[7] and could not be detected by fluorescent quantitative-PCR[8].
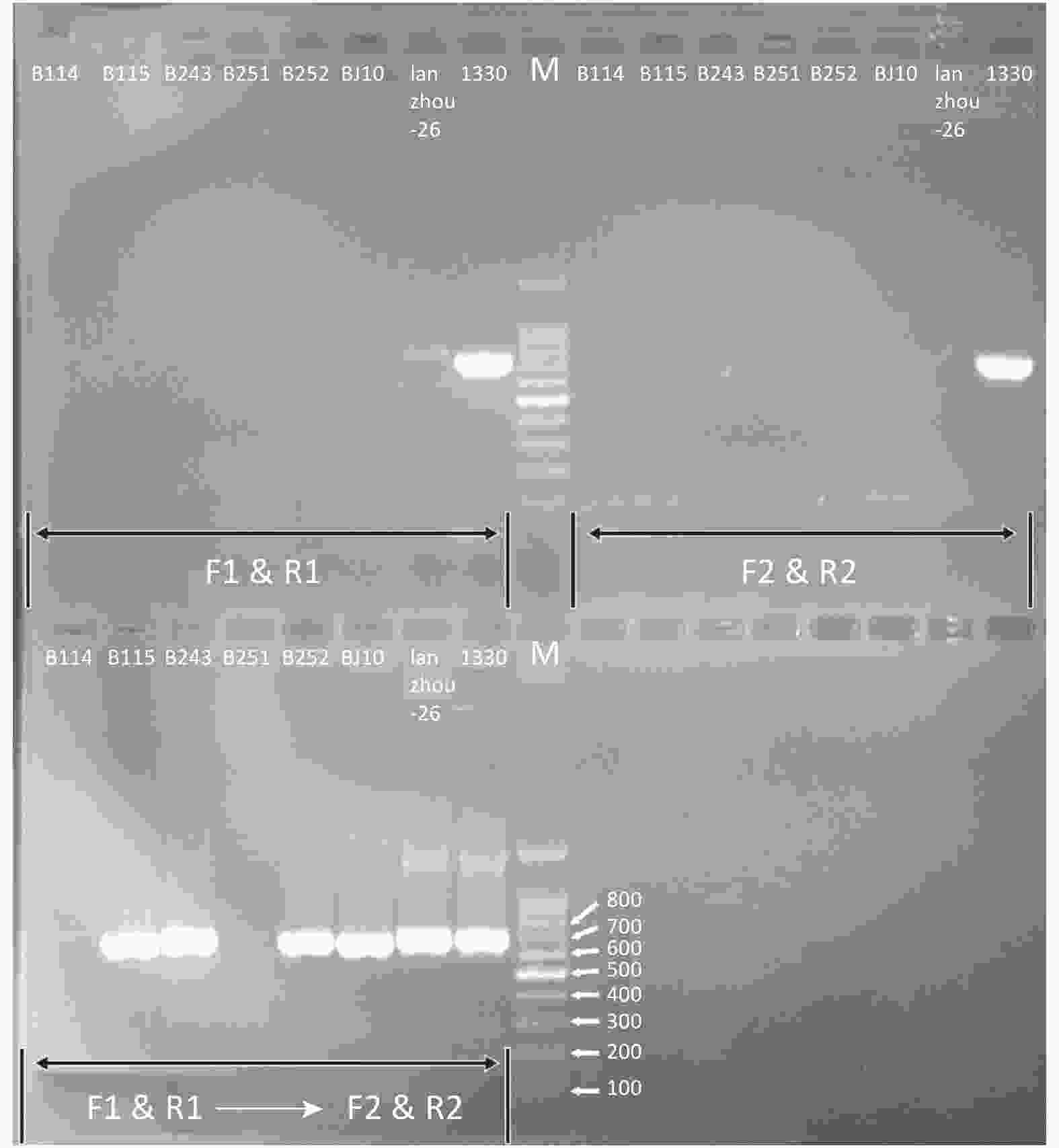
Figure 3. The nested-PCR electrophoresis of 7 extracted DNA from blood samples. The B114, B115, B243, B251, B252, BJ10, and Lanzhou-26 were the tested blood and serum samples. 1330 was Brucella suis biovars 1 strain. F1 & R1: PCR products with primers F1 and R1. F2 & R2: PCR products with primers F2 and R2. F1 & R→F2 & R2: nested-PCR products with primers F1 and R1 first and F2 and R2 second.
Recent studies have shown that PCR, either individually or multiplex reactions, can be used routinely to detect DNA from pure bacteria as they are rapid, highly specific, and sensitive. The PCR assay showed the highest sensitivity with the primer pairs B4 and B5[6], as it detected ≥ 500 CFU[7]. Due to the effects of inherent inhibitory factors in the blood and other tissues, real-time PCR assay was not deemed suitable for the detection of Brucella DNA[7]. Therefore, there is an urgent need to develop a simple and sensitive method for the identification of Brucella, especially for pathogenic bacteria in the blood and other tissue samples.
In this study, nested-PCR assay was unaffected by the inhibitory factors, and its detection sensitivity reached one copy number of Brucella DNA. Meanwhile, the species and biovars of infectious Brucella could be identified and typed. Until date, there exists no method that can completely match the biological typing for Brucella. owing to the high consistency of Brucella DNA, such as the recently proposed and widely used multiple locus variable number of tandem repeats (MLVA) analysis[8]. Fortunately, nested-PCR assay can classify human pathogenic Brucella, including B. abortus, B. melitensis, B. suis, and B. canis. The results of the present study revealed that nested-PCR can be useful for the detection and typing of Brucella DNA, including that of clinical strains and blood specimens.
-
The author are sincerely grateful to all participants in this study.
doi: 10.3967/bes2021.028
-
-
Figure 2. The cluster analysis of 36 Brucella reference strains, 89 clinical isolates, and 7 blood samples by nested-PCR. The black background of the code numbers indicates blood samples (B115, B243, B252, BJ10, and Lanzhou-26) in Figure 1.
Figure 3. The nested-PCR electrophoresis of 7 extracted DNA from blood samples. The B114, B115, B243, B251, B252, BJ10, and Lanzhou-26 were the tested blood and serum samples. 1330 was Brucella suis biovars 1 strain. F1 & R1: PCR products with primers F1 and R1. F2 & R2: PCR products with primers F2 and R2. F1 & R→F2 & R2: nested-PCR products with primers F1 and R1 first and F2 and R2 second.
Table 1. The 36 reference strains, 89 clinical isolates, and 7 blood and sera samples used in this study
NO. Strain ID Biovars Note NO. Strain D Biovars Note NO. Strain D Biovars Note NO. Strain D Biovars Note 1 A13334 A1 REF 34 63/290 Ovis REF 67 13106 M3 Isolate 100 78037 M2 Isolate 2 2308 A1 REF 35 25840 Ovis REF 68 13107 M3 Isolate 101 79029 M3 Isolate 3 9-941 A1 REF 36 B2-94 Pinni REF 69 13108 M3 Isolate 102 79054 M3 Isolate 4 86/8/59 A2 REF 37 208 S1 Isolate 70 13138 S1 Isolate 103 79077 M3 Isolate 5 Tulya A3 REF 38 226 S3 Isolate 71 13139 S2 Isolate 104 79104 M3 Isolate 6 292 A4 REF 39 1048 A3 Isolate 72 13147 S1 Isolate 105 79130 M3 Isolate 7 B3196 A5 REF 40 1057 A1 Isolate 73 13160 A1 Isolate 106 80345 M3 Isolate 8 870 A6 REF 41 1148 A3 Isolate 74 13195 M3 Isolate 107 80347 M3 Isolate 9 63/75 A7 REF 42 3001 M3 Isolate 75 13248 M2 Isolate 108 80355 M2 Isolate 10 C86 A9 REF 43 5017 M3 Isolate 76 13331 S2 Isolate 109 80356 M2 Isolate 11 104M A Vacci 44 7033 M3 Isolate 77 13332 S1 Isolate 110 80359 A3 Isolate 12 S19 A Vacci 45 8257 A1 Isolate 78 13341 M3 Isolate 111 80392 A9 Isolate 13 45/20 A REF 46 8416 A9 Isolate 79 23385 Canis Isolate 112 81006 M3 Isolate 14 B1119 A REF 47 9057 S3 Isolate 80 52141 Canis Isolate 113 81011 A7 Isolate 15 16M M1 REF 48 10009 S3 Isolate 81 57002 A3 Isolate 114 81033 M3 Isolate 16 63/9 M2 REF 49 10036 Canis Isolate 82 60033 A1 Isolate 115 84001 M3 Isolate 17 Eher M3 REF 50 10062 S3 Isolate 83 61005 M3 Isolate 116 86028 M1 Isolate 18 NI M REF 51 11038 Canis Isolate 84 63006 M2 Isolate 117 86028 S1 Isolate 19 M5 M Vacci 52 11055 A3 Isolate 85 63031 A1 Isolate 118 88040 S1 Isolate 20 M28 M Vacci 53 11059 M3 Isolate 86 64008 S3 Isolate 119 90001 A3 Isolate 21 Rev1 M Vacci 54 12088 M3 Isolate 87 64027 M2 Isolate 120 92008 M3 Isolate 22 1330 S1 REF 55 12099 M3 Isolate 88 64029 A1 Isolate 121 B257 A1 Isolate 23 Thomsen S2 REF 56 12110 M3 Isolate 89 65050 M2 Isolate 122 HN-13 M1 Isolate 24 686 S3 REF 57 13031 M3 Isolate 90 65066 M2 Isolate 123 HNLG M3 Isolate 25 40 S4 REF 58 13034 M3 Isolate 91 65079 A1 Isolate 124 HNLR M3 Isolate 26 513 S5 REF 59 13051 M3 Isolate 92 66148 M2 Isolate 125 HNMB M3 Isolate 27 VacciS2 S Vacci 60 13067 M3 Isolate 93 72003 M2 Isolate 126 B114 / Blood 28 VBI22 S Vacci 61 13080 M3 Isolate 94 73011 M3 Isolate 127 B115 A1 Blood 29 RM6/66 Canis REF 62 13081 M3 Isolate 95 73096 S3 Isolate 128 BJ10 Canis Blood 30 10759 Ceti REF 63 13083 M3 Isolate 96 73099 M3 Isolate 129 Lanzhou-26 A1 Blood 31 28753 Ceti REF 64 13096 S3 Isolate 97 73101 M3 Isolate 130 B243 S3 Serum 32 4915 Micro REF 65 13102 M3 Isolate 98 73162 M3 Isolate 131 B251 / Serum 33 5K33 Neoto REF 66 13105 M3 Isolate 99 78014 M3 Isolate 132 B252 M3 Serum Note. A: B. abortus biovar, M: B. melitensis biovar, S: B. suis biovar, Canis: B. canis, Ceti: B. ceti, Pinni: B. pinnipedialis, Neoto: B. neotomae, Ovis: B. ovis, Micro: B. microti, REF: Reference strain, Isolate: Clinical isolate. /: Brucella was not detected in the blood samples by culturing or nested-PCR assay. REF, reference. -
[1] Franco MP, Mulder M, Gilman RH, et al. Human brucellosis. Lancet Infect Dis, 2007; 7, 775−86. doi: 10.1016/S1473-3099(07)70286-4 [2] Araj GF. Update on laboratory diagnosis of human brucellosis. Int J Antimicrob Agents, 2010; 36, 12−7. doi: 10.1016/j.ijantimicag.2010.06.014 [3] Bauer S, Aubert AC, Richli M, et al. Blood cultures in the evaluation of uncomplicated cellulitis. Eur J Intern Med, 2016; 36, 50−6. doi: 10.1016/j.ejim.2016.07.029 [4] Pabuccuoglu O, Ecemis T, El S, et al. Evaluation of serological tests for diagnosis of brucellosis. Jpn J Infect Dis, 2011; 64, 272−6. [5] Tian GZ, Zheng M, Cui BY, et al. Analysis of pathogens and genetic characteristics of Brucella canis. Chin Endemiol, 2016; 35, 707−12. [6] Sidor IF, Dunn JL, Tsongalis GJ, et al. A multiplex real-time polymerase chain reaction assay with two internal controls for the detection of Brucella species in tissues, blood, and feces from marine mammals. J Vet Diagn Invest, 2013; 25, 72−81. doi: 10.1177/1040638712470945 [7] Baily GG, Krahn JB, Drasar BS, et al. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg, 1992; 95, 271−5. [8] Tian GZ, Cui BY, Piao DR, et al. Multi-locus variable-number tandem repeat analysis of Chinese Brucella strains isolated from 1953 to 2013. Infect Dis Poverty, 2017; 6, 89. doi: 10.1186/s40249-017-0296-0 -




 下载:
下载:




 Quick Links
Quick Links