-
The new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading worldwide with the number of confirmed cases increasing dramatically[1, 2]. Coronaviruses have the largest genome among RNA viruses and have caused two major outbreaks[3, 4]. The new coronavirus (SARS-CoV-2) is the third coronavirus spread globally in the past 17 years. According to the World Health Organization, SARS-CoV-2 has affected more than 155.6 millon patients in 222 countries as of 6 May 2021 and has become a major global health concern (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
Rapid genome sequencing of the virus directly from clinical and environmental specimens is important for real-time genomic surveillance in managing virus outbreaks[5]. Genomic sequencing directly from clinical and environmental samples is challenging owing to low sensitivity. Therefore, we designed a multiplex PCR method for virus genome sequencing of SARS-CoV-2 directly from clinical and environmental samples with reference to the whole-genome sequencing method for Zika virus[6]. SARS-CoV-2 can be detected in the bronchoalveolar lavage fluid, sputum, nasal swab, throat swab, feces, blood, and saliva[7, 8]. Hence, in this study, we validated the method with different types of these samples.
The complete protocol is schematically represented in Figure 1. The first step of the protocol was to design overlapping primer pairs to cover the entire genome (Supplementary Table S1 available in www.besjournal.com). The multiplex PCR comprised a set of 102 oligonucleotide primer pairs and the amplicons generated by the primer pairs spanned the target genome (Figure 1A). The subsequent step was the amplification and sequencing of fragments that were performed with Nanopore GridION X5 to obtain reads of 400 bp or Illumina paired-end library protocol, allowing reads of approximately 300 bp. During genome sequencing, some amplicons were over-sequenced, while others were under-sequenced (Figure 2). This occurs owing to differences in the amplification process and genome content.
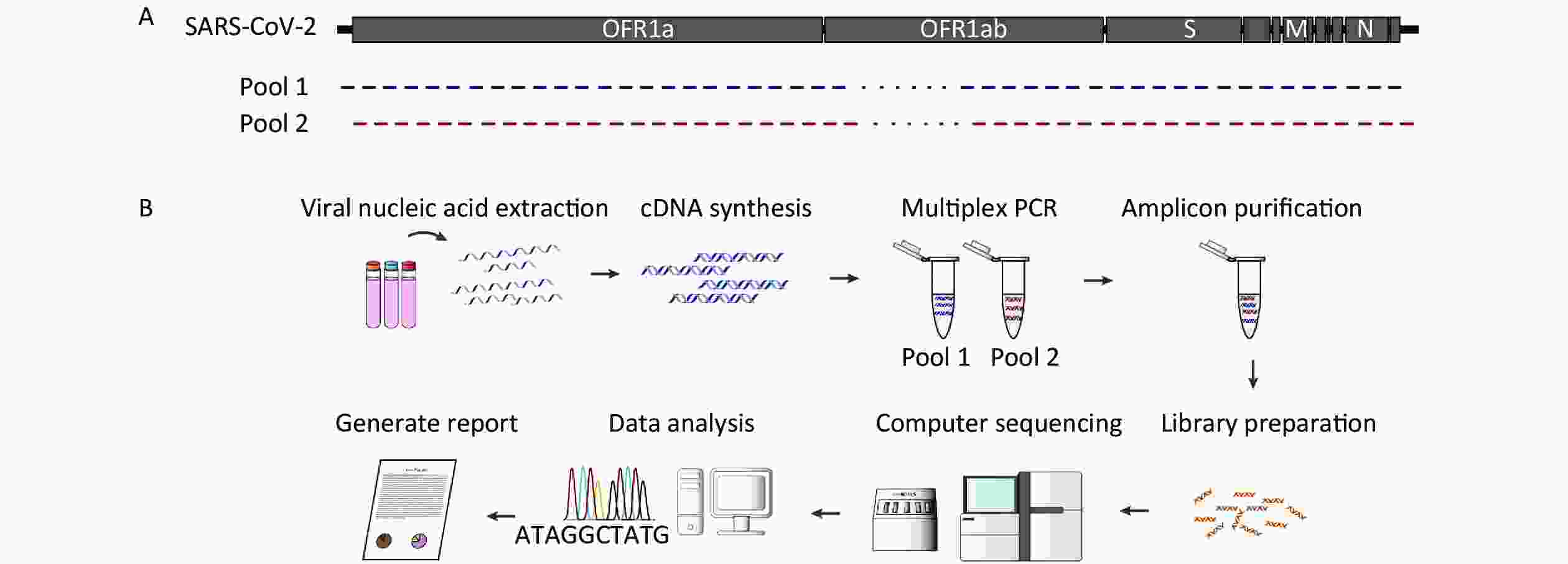
Figure 1. Sequencing schemes employed in the study. (A) Schematic showing expected amplicon products for each pool of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome. We designed and optimized polymerase chain reaction (PCR) primers to generate amplicons that would span the SARS-CoV-2 genome. We designed 102 primer pairs, which covered 100% of the SARS-CoV-2 genome. The predicted forward primers (blue arrows) and reverse primers (red arrows) scaled according to the SARS-CoV-2 virus coordinates. (B) Workflow of the multiplex PCR method for sequencing SARS-CoV-2 directly from clinical specimens.
Table S1. The primer sequences of SARS-CoV-2 genome
Primer name Pool 1/2 Sequences (5'→3') SARS-CoV-2_1_Forward Pool_1 ATTAAAGGTTTATACCTTCCCAGGTA SARS-CoV-2_1_Reverse Pool_1 CAGCCACACAGATTTTAAAGTTCGT SARS-CoV-2_2_Forward Pool_2 CCAGGTAACAAACCAACCAACTT SARS-CoV-2_2_Reverse Pool_2 GCCACACAGATTTTAAAGTTCGTTT SARS-CoV-2_3_Forward Pool_1 ACCAACCAACTTTCGATCTCTTGT SARS-CoV-2_3_Reverse Pool_1 CATCTTTAAGATGTTGACGTGCCTC SARS-CoV-2_4_Forward Pool_2 CTGTTTTACAGGTTCGCGACGT SARS-CoV-2_4_Reverse Pool_2 TAAGGATCAGTGCCAAGCTCGT SARS-CoV-2_5_Forward Pool_1 CGGTAATAAAGGAGCTGGTGGC SARS-CoV-2_5_Reverse Pool_1 AAGGTGTCTGCAATTCATAGCTCT SARS-CoV-2_6_Forward Pool_2 GGTGTATACTGCTGCCGTGAAC SARS-CoV-2_6_Reverse Pool_2 CACAAGTAGTGGCACCTTCTTTAGT SARS-CoV-2_7_Forward Pool_1 TGGTGAAACTTCATGGCAGACG SARS-CoV-2_7_Reverse Pool_1 ATTGATGTTGACTTTCTCTTTTTGGAGT SARS-CoV-2_8_Forward Pool_2 GGTGTTGTTGGAGAAGGTTCCG SARS-CoV-2_8_Reverse Pool_2 TAGCGGCCTTCTGTAAAACACG SARS-CoV-2_9_Forward Pool_1 ATCAGAGGCTGCTCGTGTTGTA SARS-CoV-2_9_Reverse Pool_1 TGCACAGGTGACAATTTGTCCA SARS-CoV-2_10_Forward Pool_2 AGAGTTTCTTAGAGACGGTTGGGA SARS-CoV-2_10_Reverse Pool_2 GCTTCAACAGCTTCACTAGTAGGT SARS-CoV-2_11_Forward Pool_1 ACTGGTGATTTACAACCATTAGAACAA SARS-CoV-2_11_Reverse Pool_1 CACAGGCGAACTCATTTACTTCTGTA SARS-CoV-2_12_Forward Pool_2 TGAGAAGTGCTCTGCCTATACAGT SARS-CoV-2_12_Reverse Pool_2 TCATCTAACCAATCTTCTTCTTGCTCT SARS-CoV-2_13_Forward Pool_1 GGAATTTGGTGCCACTTCTGCT SARS-CoV-2_13_Reverse Pool_1 TCATCAGATTCAACTTGCATGGCA SARS-CoV-2_14_Forward Pool_2 AAACATGGAGGAGGTGTTGCAG SARS-CoV-2_14_Reverse Pool_2 TTCACTCTTCATTTCCAAAAAGCTTGA SARS-CoV-2_15_Forward Pool_1 TCGCACAAATGTCTACTTAGCTGT SARS-CoV-2_15_Reverse Pool_1 ACCACAGCAGTTAAAACACCCT SARS-CoV-2_16_Forward Pool_2 CATCCAGATTCTGCCACTCTTGT SARS-CoV-2_16_Reverse Pool_2 AGTTTCCACACAGACAGGCATT SARS-CoV-2_17_Forward Pool_1 ACAGTGCTTAAAAAGTGTAAAAGTGCC SARS-CoV-2_17_Reverse Pool_1 AACAGAAACTGTAGCTGGCACT SARS-CoV-2_18_Forward Pool_2 AATTTGGAAGAAGCTGCTCGGT SARS-CoV-2_18_Reverse Pool_2 CACAACTTGCGTGTGGAGGTTA SARS-CoV-2_19_Forward Pool_1 CTTCTTTCTTTGAGAGAAGTGAGGACT SARS-CoV-2_19_Reverse Pool_1 TTTGTTGGAGTGTTAACAATGCAGT SARS-CoV-2_20_Forward Pool_2 ACAACTGTTATCTTGCCACTGCAT SARS-CoV-2_20_Reverse Pool_2 AAATTGTTCATAAGAAAGTGTGCCC SARS-CoV-2_21_Forward Pool_1 GCTGTTATGTACATGGGCACACT SARS-CoV-2_21_Reverse Pool_1 TGTCCAACTTAGGGTCAATTTCTGT SARS-CoV-2_22_Forward Pool_2 ACAAAGAAAACAGTTACACAACAACCA SARS-CoV-2_22_Reverse Pool_2 ACGTGGCTTTATTAGTTGCATTGTT SARS-CoV-2_23_Forward Pool_1 TGGCTATTGATTATAAACACTACACACCC SARS-CoV-2_23_Reverse Pool_1 TAGATCTGTGTGGCCAACCTCT SARS-CoV-2_24_Forward Pool_2 ACTACCGAAGTTGTAGGAGACATTATACT SARS-CoV-2_24_Reverse Pool_2 ACAGTATTCTTTGCTATAGTAGTCGGC SARS-CoV-2_25_Forward Pool_1 ACAACTACTAACATAGTTACACGGTGT SARS-CoV-2_25_Reverse Pool_1 ACCAGTACAGTAGGTTGCAATAGTG SARS-CoV-2_26_Forward Pool_2 AGGCATGCCTTCTTACTGTACTG SARS-CoV-2_26_Reverse Pool_2 ACATTCTAACCATAGCTGAAATCGGG SARS-CoV-2_27_Forward Pool_1 GCAATTGTTTTTCAGCTATTTTGCAGT SARS-CoV-2_27_Reverse Pool_1 ACTGTAGTGACAAGTCTCTCGCA SARS-CoV-2_28_Forward Pool_2 TTGTGATACATTCTGTGCTGGTAGT SARS-CoV-2_28_Reverse Pool_2 TCCGCACTATCACCAACATCAG SARS-CoV-2_29_Forward Pool_1 ACTACAGTCAGCTTATGTGTCAACC SARS-CoV-2_29_Reverse Pool_1 AATACAAGCACCAAGGTCACGG SARS-CoV-2_30_Forward Pool_2 ACATAGAAGTTACTGGCGATAGTTGT SARS-CoV-2_30_Reverse Pool_2 TGTTTAGACATGACATGAACAGGTGT SARS-CoV-2_31_Forward Pool_1 ACTTGTGTTCCTTTTTGTTGCTGC SARS-CoV-2_31_Reverse Pool_1 AGTGTACTCTATAAGTTTTGATGGTGTGT SARS-CoV-2_32_Forward Pool_2 GCACAACTAATGGTGACTTTTTGCA SARS-CoV-2_32_Reverse Pool_2 ACCACTAGTAGATACACAAACACCAG SARS-CoV-2_33_Forward Pool_1 TTCTGAGTACTGTAGGCACGGC SARS-CoV-2_33_Reverse Pool_1 ACAGAATAAACACCAGGTAAGAATGAGT SARS-CoV-2_34_Forward Pool_2 TGGTGAATACAGTCATGTAGTTGCC SARS-CoV-2_34_Reverse Pool_2 AGCACATCACTACGCAACTTTAGA SARS-CoV-2_35_Forward Pool_1 ACTTTTGAAGAAGCTGCGCTGT SARS-CoV-2_35_Reverse Pool_1 TGGACAGTAAACTACGTCATCAAGC SARS-CoV-2_36_Forward Pool_2 TCCCATCTGGTAAAGTTGAGGGT SARS-CoV-2_36_Reverse Pool_2 AGTGAAATTGGGCCTCATAGCA SARS-CoV-2_37_Forward Pool_1 TGTTCGCATTCAACCAGGACAG SARS-CoV-2_37_Reverse Pool_1 ACTTCATAGCCACAAGGTTAAAGTCA SARS-CoV-2_38_Forward Pool_2 TTAGCTTGGTTGTACGCTGCTG SARS-CoV-2_38_Reverse Pool_2 GAACAAAGACCATTGAGTACTCTGGA SARS-CoV-2_39_Forward Pool_1 ACACACCACTGGTTGTTACTCAC SARS-CoV-2_39_Reverse Pool_1 GTCCACACTCTCCTAGCACCAT SARS-CoV-2_40_Forward Pool_2 ACTGTGTTATGTATGCATCAGCTGT SARS-CoV-2_40_Reverse Pool_2 CACCAAGAGTCAGTCTAAAGTAGCG SARS-CoV-2_41_Forward Pool_1 AGTATTGCCCTATTTTCTTCATAACTGGT SARS-CoV-2_41_Reverse Pool_1 TGTAACTGGACACATTGAGCCC SARS-CoV-2_42_Forward Pool_2 TGCACATCAGTAGTCTTACTCTCAGT SARS-CoV-2_42_Reverse Pool_2 CATGGCTGCATCACGGTCAAAT SARS-CoV-2_43_Forward Pool_1 GTTCCCTTCCATCATATGCAGCT SARS-CoV-2_43_Reverse Pool_1 TGGTATGACAACCATTAGTTTGGCT SARS-CoV-2_44_Forward Pool_2 TGCAAGAGATGGTTGTGTTCCC SARS-CoV-2_44_Reverse Pool_2 CCTACCTCCCTTTGTTGTGTTGT SARS-CoV-2_45_Forward Pool_1 TACGACAGATGTCTTGTGCTGC SARS-CoV-2_45_Reverse Pool_1 AGCAGCATCTACAGCAAAAGCA SARS-CoV-2_46_Forward Pool_2 TGCCACAGTACGTCTACAAGCT SARS-CoV-2_46_Reverse Pool_2 AACCTTTCCACATACCGCAGAC SARS-CoV-2_47_Forward Pool_1 CCTGTGGGTTTTACACTTAAAAACA SARS-CoV-2_47_Reverse Pool_1 AATTGTTTCTTCATGTTGGTAGTTAGAG SARS-CoV-2_48_Forward Pool_2 TGTCGCTTCCAAGAAAAGGACG SARS-CoV-2_48_Reverse Pool_2 CACGTTCACCTAAGTTGGCGTA SARS-CoV-2_49_Forward Pool_1 AGGACTGGTATGATTTTGTAGAAAACCC SARS-CoV-2_49_Reverse Pool_1 AATAACGGTCAAAGAGTTTTAACCTCTC SARS-CoV-2_50_Forward Pool_2 TGTTGACACTGACTTAACAAAGCCT SARS-CoV-2_50_Reverse Pool_2 TAGATTACCAGAAGCAGCGTGC SARS-CoV-2_51_Forward Pool_1 AGGAATTACTTGTGTATGCTGCTGA SARS-CoV-2_51_Reverse Pool_1 TGACGATGACTTGGTTAGCATTAATACA SARS-CoV-2_52_Forward Pool_2 GTTGATAAGTACTTTGATTGTTACGATGGT SARS-CoV-2_52_Reverse Pool_2 TAACATGTTGTGCCAACCACCA SARS-CoV-2_53_Forward Pool_1 TCAATAGCCGCCACTAGAGGAG SARS-CoV-2_53_Reverse Pool_1 AGTGCATTAACATTGGCCGTGA SARS-CoV-2_54_Forward Pool_2 CATCAGGAGATGCCACAACTGC SARS-CoV-2_54_Reverse Pool_2 GTTGAGAGCAAAATTCATGAGGTCC SARS-CoV-2_55_Forward Pool_1 AGCAAAATGTTGGACTGAGACTGA SARS-CoV-2_55_Reverse Pool_1 AGCCTCATAAAACTCAGGTTCCC SARS-CoV-2_56_Forward Pool_2 TGAGTTAACAGGACACATGTTAGACA SARS-CoV-2_56_Reverse Pool_2 AACCAAAAACTTGTCCATTAGCACA SARS-CoV-2_57_Forward Pool_1 ACTCAACTTTACTTAGGAGGTATGAGCT SARS-CoV-2_57_Reverse Pool_1 GGTGTACTCTCCTATTTGTACTTTACTGT SARS-CoV-2_58_Forward Pool_2 ACCTAGACCACCACTTAACCGA SARS-CoV-2_58_Reverse Pool_2 ACACTATGCGAGCAGAAGGGTA SARS-CoV-2_59_Forward Pool_1 ATTCTACACTCCAGGGACCACC SARS-CoV-2_59_Reverse Pool_1 GTAATTGAGCAGGGTCGCCAAT SARS-CoV-2_60_Forward Pool_2 TGATTTGAGTGTTGTCAATGCCAGA SARS-CoV-2_60_Reverse Pool_2 CTTTTCTCCAAGCAGGGTTACGT SARS-CoV-2_61_Forward Pool_1 TCACGCATGATGTTTCATCTGCA SARS-CoV-2_61_Reverse Pool_1 AAGAGTCCTGTTACATTTTCAGCTTG SARS-CoV-2_62_Forward Pool_2 TGATAGAGACCTTTATGACAAGTTGCA SARS-CoV-2_62_Reverse Pool_2 GGTACCAACAGCTTCTCTAGTAGC SARS-CoV-2_63_Forward Pool_1 TGTTTATCACCCGCGAAGAAGC SARS-CoV-2_63_Reverse Pool_1 ATCACATAGACAACAGGTGCGC SARS-CoV-2_64_Forward Pool_2 GGCACATGGCTTTGAGTTGACA SARS-CoV-2_64_Reverse Pool_2 GTTGAACCTTTCTACAAGCCGC SARS-CoV-2_65_Forward Pool_1 TGTTAAGCGTGTTGACTGGACT SARS-CoV-2_65_Reverse Pool_1 ACAAACTGCCACCATCACAACC SARS-CoV-2_66_Forward Pool_2 TCGATAGATATCCTGCTAATTCCATTGT SARS-CoV-2_66_Reverse Pool_2 AGTCTTGTAAAAGTGTTCCAGAGGT SARS-CoV-2_67_Forward Pool_1 GCTGGCTTTAGCTTGTGGGTTT SARS-CoV-2_67_Reverse Pool_1 TGTCAGTCATAGAACAAACACCAATAGT SARS-CoV-2_68_Forward Pool_2 GGGTGTGGACATTGCTGCTAAT SARS-CoV-2_68_Reverse Pool_2 TCAATTTCCATTTGACTCCTGGGT SARS-CoV-2_69_Forward Pool_1 GTTGTCCAACAATTACCTGAAACTTACT SARS-CoV-2_69_Reverse Pool_1 CAACCTTAGAAACTACAGATAAATCTTGGG SARS-CoV-2_70_Forward Pool_2 ACAGGTTCATCTAAGTGTGTGTGT SARS-CoV-2_70_Reverse Pool_2 CTCCTTTATCAGAACCAGCACCA SARS-CoV-2_71_Forward Pool_1 TGTCGCAAAATATACTCAACTGTGTCA SARS-CoV-2_71_Reverse Pool_1 TCTTTATAGCCACGGAACCTCCA SARS-CoV-2_72_Forward Pool_2 ACAAAAGAAAATGACTCTAAAGAGGGTTT SARS-CoV-2_72_Reverse Pool_2 TGACCTTCTTTTAAAGACATAACAGCAG SARS-CoV-2_73_Forward Pool_1 ACAAATCCAATTCAGTTGTCTTCCTATTC SARS-CoV-2_73_Reverse Pool_1 TGGAAAAGAAAGGTAAGAACAAGTCCT SARS-CoV-2_74_Forward Pool_2 ACACGTGGTGTTTATTACCCTGAC SARS-CoV-2_74_Reverse Pool_2 ACTCTGAACTCACTTTCCATCCAAC SARS-CoV-2_75_Forward Pool_1 CAATTTTGTAATGATCCATTTTTGGGTGT SARS-CoV-2_75_Reverse Pool_1 CACCAGCTGTCCAACCTGAAGA SARS-CoV-2_76_Forward Pool_2 ACATCACTAGGTTTCAAACTTTACTTGC SARS-CoV-2_76_Reverse Pool_2 GCAACACAGTTGCTGATTCTCTTC SARS-CoV-2_77_Forward Pool_1 AGAGTCCAACCAACAGAATCTATTGT SARS-CoV-2_77_Reverse Pool_1 ACCACCAACCTTAGAATCAAGATTGT SARS-CoV-2_78_Forward Pool_2 GGCAAACTGGAAAGATTGCTGA SARS-CoV-2_78_Reverse Pool_2 TTGAAATTGACACATTTGTTTTTAACC SARS-CoV-2_79_Forward Pool_1 CCAGCAACTGTTTGTGGACCTA SARS-CoV-2_79_Reverse Pool_1 CAGCCCCTATTAAACAGCCTGC SARS-CoV-2_80_Forward Pool_2 CAACTTACTCCTACTTGGCGTGT SARS-CoV-2_80_Reverse Pool_2 TGTGTACAAAAACTGCCATATTGCA SARS-CoV-2_81_Forward Pool_1 GTGGTGATTCAACTGAATGCAGC SARS-CoV-2_81_Reverse Pool_1 CATTTCATCTGTGAGCAAAGGTGG SARS-CoV-2_82_Forward Pool_2 TTGCCTTGGTGATATTGCTGCT SARS-CoV-2_82_Reverse Pool_2 TGGAGCTAAGTTGTTTAACAAGCG SARS-CoV-2_83_Forward Pool_1 GCACTTGGAAAACTTCAAGATGTGG SARS-CoV-2_83_Reverse Pool_1 GTGAAGTTCTTTTCTTGTGCAGGG SARS-CoV-2_84_Forward Pool_2 GGGCTATCATCTTATGTCCTTCCCT SARS-CoV-2_84_Reverse Pool_2 TGCCAGAGATGTCACCTAAATCAA SARS-CoV-2_85_Forward Pool_1 TCCTTTGCAACCTGAATTAGACTCA SARS-CoV-2_85_Reverse Pool_1 TTTGACTCCTTTGAGCACTGGC SARS-CoV-2_86_Forward Pool_2 TGCTGTAGTTGTCTCAAGGGCT SARS-CoV-2_86_Reverse Pool_2 AGGTGTGAGTAAACTGTTACAAACAAC SARS-CoV-2_87_Forward Pool_1 ACTAGCACTCTCCAAGGGTGTT SARS-CoV-2_87_Reverse Pool_1 ACACAGTCTTTTACTCCAGATTCCC SARS-CoV-2_88_Forward Pool_2 TCAGGTGATGGCACAACAAGTC SARS-CoV-2_88_Reverse Pool_2 ACGAAAGCAAGAAAAAGAAGTACGC SARS-CoV-2_89_Forward Pool_1 CGACTACTAGCGTGCCTTTGTA SARS-CoV-2_89_Reverse Pool_1 ACTAGGTTCCATTGTTCAAGGAGC SARS-CoV-2_90_Forward Pool_2 CCATGGCAGATTCCAACGGTAC SARS-CoV-2_90_Reverse Pool_2 TGGTCAGAATAGTGCCATGGAGT SARS-CoV-2_91_Forward Pool_1 TCTTGTAGGCTTGATGTGGCT SARS-CoV-2_91_Reverse Pool_1 TGCTACTGGAATGGTCTGTGTTTA SARS-CoV-2_92_Forward Pool_2 ACACAGACCATTCCAGTAGCAGT SARS-CoV-2_92_Reverse Pool_2 TGAAATGGTGAATTGCCCTCGT SARS-CoV-2_93_Forward Pool_1 TCACTACCAAGAGTGTGTTAGAGGT SARS-CoV-2_93_Reverse Pool_1 TTCAAGTGAGAACCAAAAGATAATAAGCA SARS-CoV-2_94_Forward Pool_2 TTTGTGCTTTTTAGCCTTTCTGCT SARS-CoV-2_94_Reverse Pool_2 AGGTTCCTGGCAATTAATTGTAAAAGG SARS-CoV-2_95_Forward Pool_1 TGAGGCTGGTTCTAAATCACCCA SARS-CoV-2_95_Reverse Pool_1 AGGTCTTCCTTGCCATGTTGAG SARS-CoV-2_96_Forward Pool_2 GGCCCCAAGGTTTACCCAATAA SARS-CoV-2_96_Reverse Pool_2 TTTGGCAATGTTGTTCCTTGAGG SARS-CoV-2_97_Forward Pool_1 TGAGGGAGCCTTGAATACACCA SARS-CoV-2_97_Reverse Pool_1 CAGTACGTTTTTGCCGAGGCTT SARS-CoV-2_98_Forward Pool_2 GCCAACAACAACAAGGCCAAAC SARS-CoV-2_98_Reverse Pool_2 TAGGCTCTGTTGGTGGGAATGT SARS-CoV-2_99_Forward Pool_1 TGGATGACAAAGATCCAAATTTCAAAGA SARS-CoV-2_99_Reverse Pool_1 ACACACTGATTAAAGATTGCTATGTGAG SARS-CoV-2_100_Forward Pool_2 AACAATTGCAACAATCCATGAGCA SARS-CoV-2_100_Reverse Pool_2 TTCTCCTAAGAAGCTATTAAAATCACATGG SARS-CoV-2_101_Forward Pool_1 CTCACATAGCAATCTTTAATCAGTGTG SARS-CoV-2_101_Reverse Pool_1 GAGAGCTGCCTATATGGAAGAGC SARS-CoV-2_102_Forward Pool_2 TTTGTCATTCTCCTAAGAAGCTATTAA SARS-CoV-2_102_Reverse Pool_2 CCTAAGAAGCTATTAAAATCACATGGG 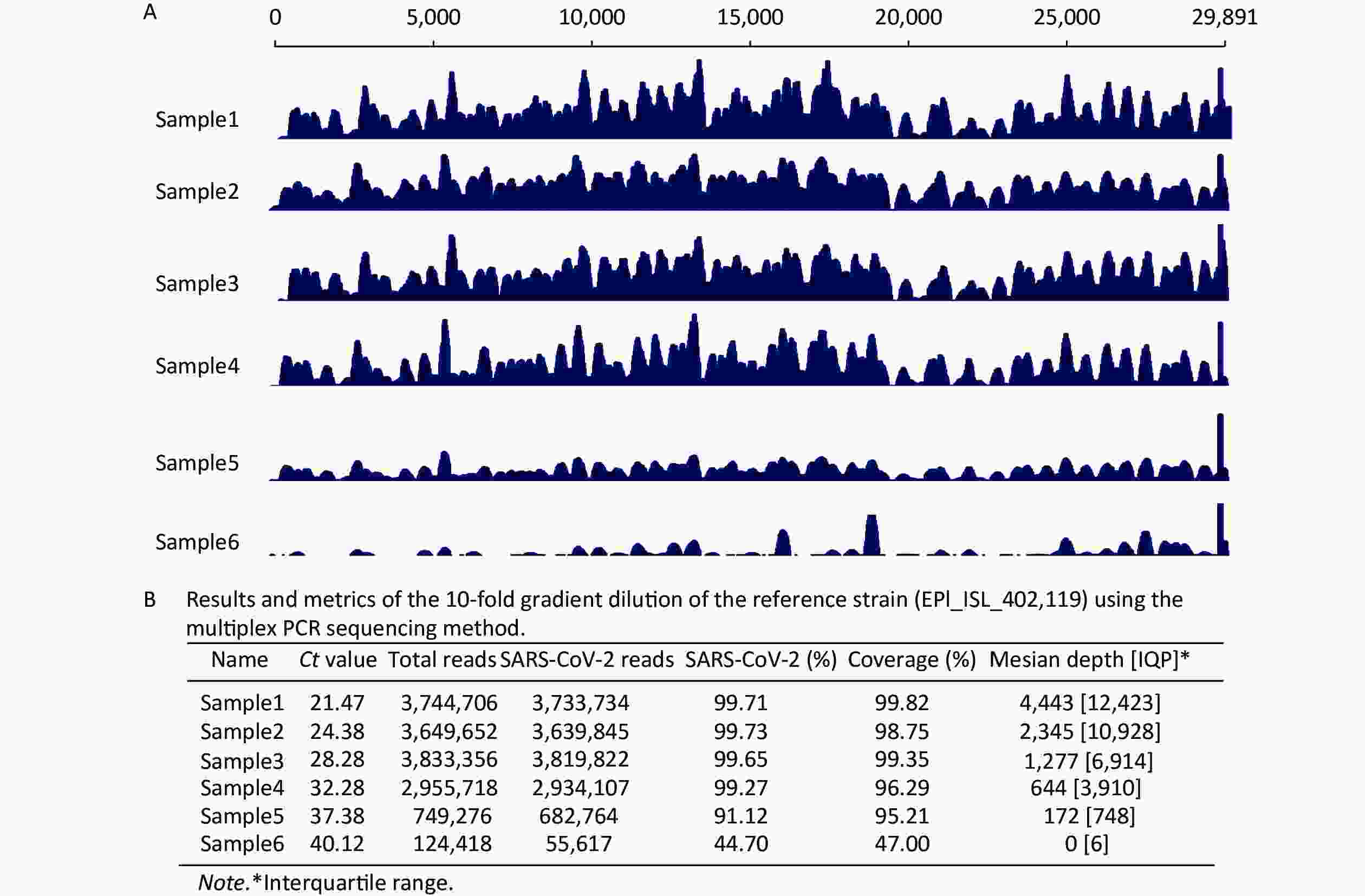
Figure 2. Study of the six diluted strains using the multiplex polymerase chain reaction sequencing method on a Nanopore GridION X5. (A) Analysis of the severe acute respiratory syndrome coronavirus 2 genome assembly at various read depths. The longest contiguous sequence produced at each read depth as a fraction of the full genome length of six diluted samples is shown. (B) Summary of statistical analysis for the sequencing results of six strains.
We evaluated the sensitivity of the multiplex PCR sequencing method using a 10-fold gradient dilution of the reference strain (EPI_ISL_402119). The sequencing results for Nanopore GridION X5 showed that the mapping reads of six gradient dilution samples against the SARS-CoV-2 reference (EPI_ISL_402119) were 3,733.734 (99.71%), 3,639.845 (99.73%), 3,819.822 (99.65%), 2,934.107 (99.27%), 682.764 (91.12%), and 55.617 (44.7%), respectively (Figure 2B). We identified that as the cycle threshold (Ct) value increased, the percentage of SARS-CoV-2 reads and the percentage of SARS-CoV-2 genome coverage showed a downward trend (Figure 2A). Furthermore, when the Ct value was < 37, the coverage of the SARS-CoV-2 genome was > 95%. Interestingly, when the Ct value was > 40, the coverage of the SARS-CoV-2 genome was < 50%. The median depth and interquartile range (IQR) of coverage of all samples are listed in Figure 2B, whereas the results on the Illumina MiSeq platform are not presented.
In this study, we used 14 different types of clinical and environmental specimens, including bronchoalveolar lavage fluid, sputum, nasal swabs, throat swabs, and feces samples, from patients infected with the new coronavirus and environmental samples. An in-depth summary, including detailed information on sequence reads, depth distributions, and genome coverage per sample, of the outputs from all samples is presented in Table 1. In comparison with previous studies and despite the long genome of the virus, sequencing of the SARS-CoV-2 genome was very specific. The number of reads that mapped against the SARS-CoV-2 sequence was very high (> 96% for 10 of the samples), revealing that the primer design and the biological protocol led to some noise in the sequencing data. A total of 11 full-length or near-full-length SARS-CoV-2 genomes (> 99% genome coverage) were obtained from 14 libraries.
Table 1. Results of amplicon scheme sequencing on fourteen SARS-CoV-2 positive clinical and environmental samples in China using the multiplex PCR sequencing method on Nanopore GridION X5
Sample name Sample type Ct value Total reads SARS-CoV-2 reads SARS-CoV-2 (%) Coverage (%) Median depth [IQR]a hCoV-19_Sample1 Alveolar lavage fluid 27.43 1,272,772 1,255,249 98.62 99.85 5,298 [4,701] hCoV-19_Sample2 Alveolar lavage fluid 31.71 1,789,508 1,735,201 96.97 99.83 5,951 [6,161] hCoV-19_Sample3 Sputum 38.00 598,376 399,343 66.74 77.15 30 [1,041] hCoV-19_Sample4 Sputum 19.06 1,865,190 1,837,254 98.50 99.81 5,492 [9,965] hCoV-19_Sample5 Sputum 25.14 2,678,180 2,628,756 98.15 99.87 8,832 [6,412] hCoV-19_Sample6 Nasal swabs 33.25 1,279,020 1,233,595 96.45 99.82 4,563 [5,926] hCoV-19_Sample7 Throat swabs 30.68 2,031,002 1,982,592 97.62 99.83 7,609 [8,081] hCoV-19_Sample8 Throat swabs 22.77 1,912,014 1,889,140 98.80 99.81 6,607 [4,459] hCoV-19_Sample9 Throat swabs 22.17 1,724,740 1,706,097 98.92 99.84 6,398 [3,722] hCoV-19_Sample10 Feces 36.44 312,000 150,863 48.35 96.57 751 [1,435] hCoV-19_Sample11 Feces 25.95 1,022,978 1,008,660 98.60 99.82 3,768 [2,382] hCoV-19_Sample12 Feces 26.06 1,771,730 1,753,368 98.96 99.83 7,078 [4,750] hCoV-19_Sample13 Environmental samples 36.06 268,000 159,279 59.43 99.69 874 [1,411] hCoV-19_Sample14 Environmental samples 36.54 312,150 150,754 48.30 98.99 732 [1,390] Note. aInterquartile range. Sequencing depth was not uniform among the amplicons along the genome; some amplicons were over-sequenced (median depth: 8,832×), while others were only sequenced a few times or not at all. Quantitative results of samples showed that all samples with relatively low Ct values (high viral load) led to a complete or nearly complete assembly. Three of the samples, hCoV-19_Sample3, hCoV-19_Sample10, and hCoV-19_Sample14, with higher Ct values (lower viral load), led to a final assembly covering only 77.15%, 96.57%, and 98.99% of the genome sequence, respectively. The significant benefit of utilizing a multiplex PCR method for virus genome sequencing over a Sanger sequencing or an unbiased metagenomic approach was the substantial increase in the number of reads specific to the viral genome[6, 9]. The number of merged reads was between 150754 and 2628756, providing sufficient sequencing data for reliable analysis of the 14 samples. We constructed a phylogenetic tree based on the full-length genome sequences of SARS-CoV-2 derived from sequencing (Supplementary Figure S1 available in www.besjournal.com).
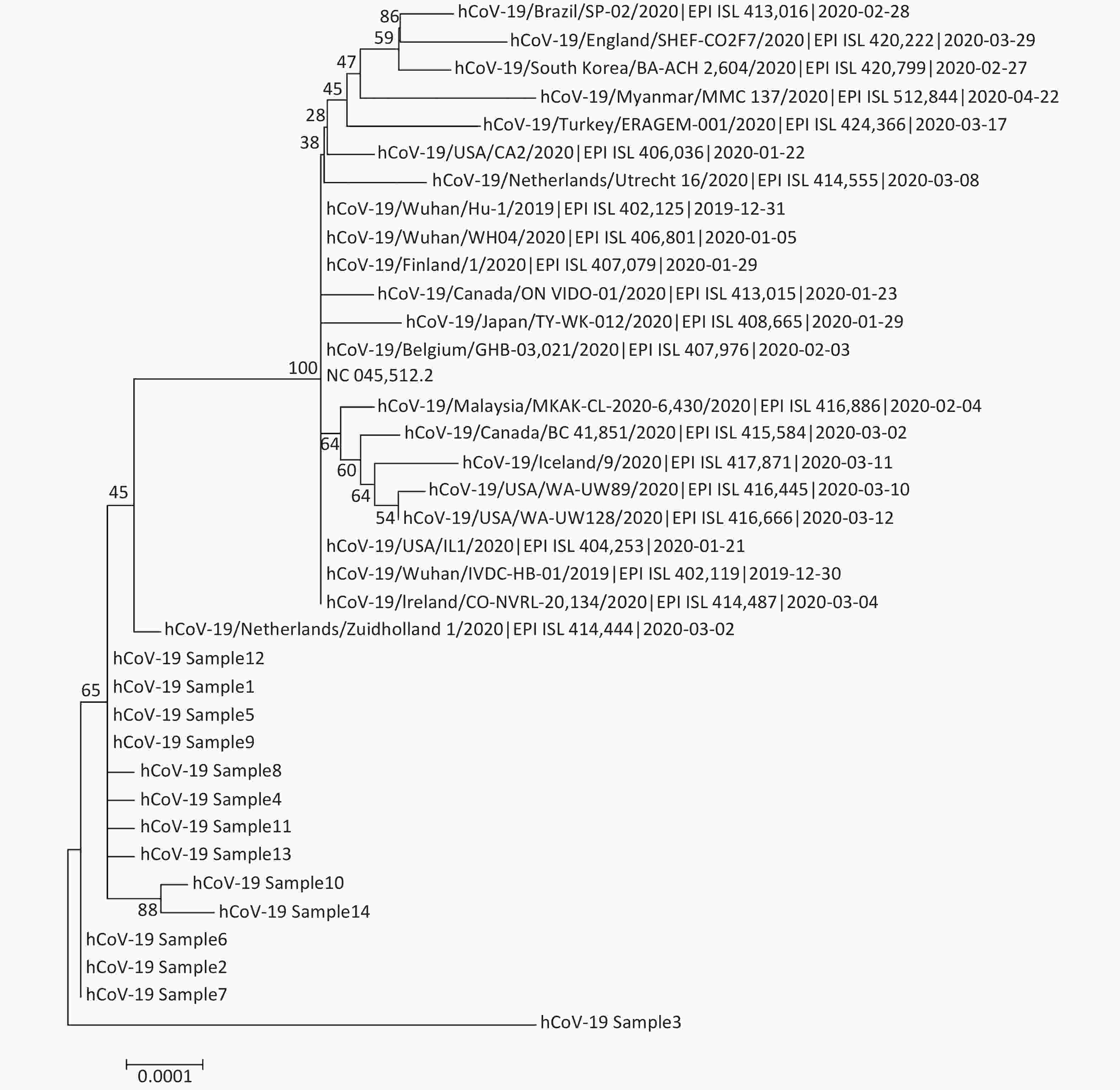
Figure S1. Phylogenetic tree based on full-length genome sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) derived by sequencing
A limitation of this study is that our method is not suitable for identifying novel viruses as primers are SARS-CoV-2 specific. Amplicon sequencing may result in incomplete genome coverage, especially when lower abundance viral genomes are present, and the loss of both 5’ and 3’ regions. To obtain complete genomes, it may be necessary to replace the problematic primers or adjust their concentration according to other primers.
In summary, our method showed advantageous prospects for generating new coronavirus sequences directly from clinical and environmental samples; however, further studies are required to confirm this finding. On a long-term basis, this method can potentially be used as a routine laboratory test to aid in treatment, vaccine design and deployment, infection control strategies, and surveillance.
全文HTML
 21084.pdf
21084.pdf
|

|



 下载:
下载:



 Quick Links
Quick Links