-
In 2019, the newly emerged SARS-CoV-2 virus caused pneumonia-like illness. The disease rapidly spread globally, leading to a worldwide outbreak referred to as the COVID-19 pandemic. The affected patients show symptoms of fever, dry cough, respiratory distress, myalgia, and gastrointestinal disturbance. As of April 5, 2021, 132,083,022 people worldwide were affected by COVID-19, while 2,868,454 people died due to the disease[1]. SARS-CoV-2-positive patients may remain asymptomatic or start showing symptoms in 2−14 days after exposure to the virus[2]. The viral infection can be diagnosed from nasopharyngeal, throat, alveolar lavage, lacrimal, blood, and stool samples. The patient starts shedding the virus in stool regardless of being symptomatic or asymptomatic, which makes sewage-based detection of the virus to be more beneficial in the early infection stage.
Raw sewage is one of the significant sources of pathogens and represents a useful matrix to study viruses excreted by humans and animals. Sewage monitoring has been previously used in different diseases by the WHO[3]. The virus has proven itself to be devastating across the world, including Pakistan, which led the Government of Pakistan to implement a lockdown since March 24, 2020. For countries with a resource-limited setting, such as Pakistan, where a large number of population relies exclusively on daily wages, masses cannot survive a complete lockdown for an extended period. With this background, in collaboration with the Water and Sanitation Agency (WASA) Lahore, we conducted the present study to predict sewage-based disease burden in a specific area or setting to facilitate government authorities to implement an effective smart lockdown or otherwise.
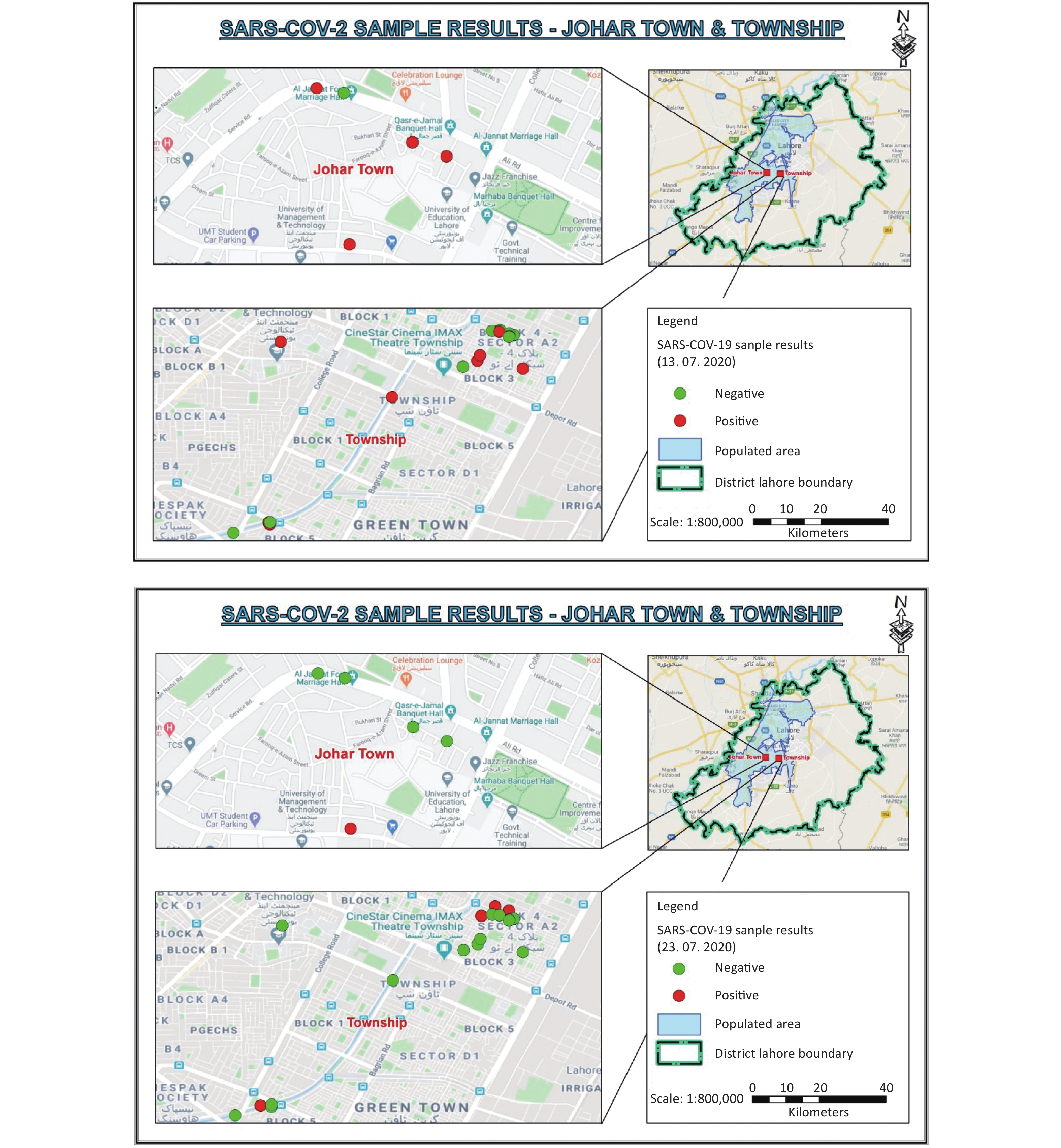
Lahore is the capital of the Punjab Province, with an estimated population of 12,642,423[4]. During the COVID-19 pandemic, the Government of Pakistan used a smart lockdown strategy to combat the spread of COVID-19. On July 9, 2020, a two-week smart lockdown was imposed in seven areas of Lahore, Pakistan, when there were 41,014 active cases. We selected two areas with smart lockdown (Johar Town C Block and Township A-II sector) for the longitudinal detection and quantification of the SARS-CoV-2 genome in sewage water of these areas during the lockdown (Figure 1). Lahore is administratively divided in 9 towns, and these lock down areas fall in Iqbal Town. There were 1,029 new COVID-19 cases in the lockdown and adjoining areas (Iqbal Town) in a period of 14 days before the start of lockdown on 9th July, 2020.
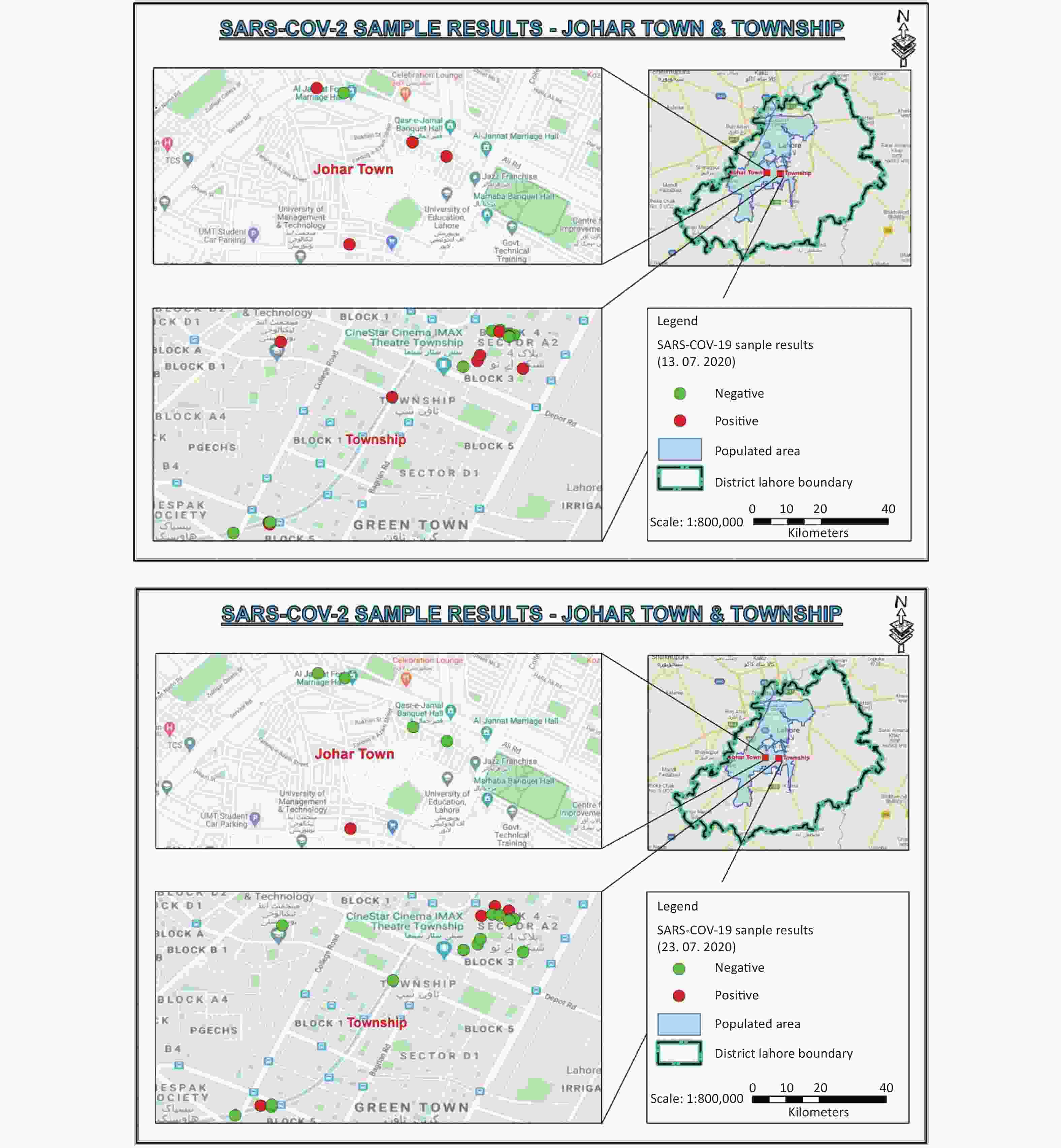
Figure 1. Satellite map of Johar Town and Township areas. Red dots indicate the SARS-CoV-2 positive samples, while green dots represent SARS-CoV-2 negative samples in the smart lockdown areas on July 13 and July 23, 2020.
Sewage water samples were collected from 5 preselected sites (4 sewerage lines and 1 lift station) of Johar Town C Block and 23 sites (19 sewerage lines and 4 lift/disposal stations) of Township A-II sector by using a grab sampling method. Sewage water samples were collected on alternate days from July 13 to July 25, 2020. Furthermore, a 24-h sewage water sampling was performed to determine the effect of sampling time on the detection and quantification of the SARS-CoV-2 genome. At the time of sewage sampling, the sampling personnel used standard personal protective equipment such as long pants, boots, hats, safety glasses, and gloves. Samples were transported to the BSL-3 facility at the Institute of Microbiology (IM), University of Veterinary and Animal Sciences (UVAS), Lahore, Pakistan, in a cool box and stored at 4 °C until further analysis within 24 h.
Viral RNA was extracted from each sewage sample in the BSL-3 facility of IM. A 200 mL sewage sample was collected by the grab sampling method and centrifuged at 3,000 ×g to remove debris and solids. After centrifugation, 1,200 µL of the supernatant was transferred to a 1.5 mL microfuge tube and spun at 5,000 ×g for 15 min at 4 °C. A 200 µL supernatant was used for viral nucleic acid extraction by using the Viral Nucleic Acid Extraction Kit II (Geneaid Biotech Ltd., Taipei, Taiwan, China)[5]. RNA was extracted using the Ascend Hero32 extraction system (Luoyang Ascend Biotech. Co. Ltd, Luoyang, China). RT-qPCR analysis of sewage samples was performed using 2019-nCoV Nucleic Acid Diagnostic Kit (Sansure Biotech Inc., Xiangya, China). We targeted the ORF1ab gene to detect SARS-CoV-2 by RT-qPCR. Thermal cycling reactions were performed at 50 °C for 30 min, followed by denaturation at 95 °C for 1 min and 45 cycles of 95 °C for 15 s and 60 °C for 30 s on the CFX-96 Real-Time PCR detection system (Bio-Rad Laboratories, Berkeley, USA). All RT-qPCR reactions had positive and negative controls. Results were considered to be positive if the cycle threshold (CT) was below 43 cycles. To calculate the copies of SARS-CoV-2 genome in sewage, a standard curve based on the ORF1ab gene was generated from different dilutions of positive control of the kit (Supplementary Figure S1, available in www.besjournal.com).
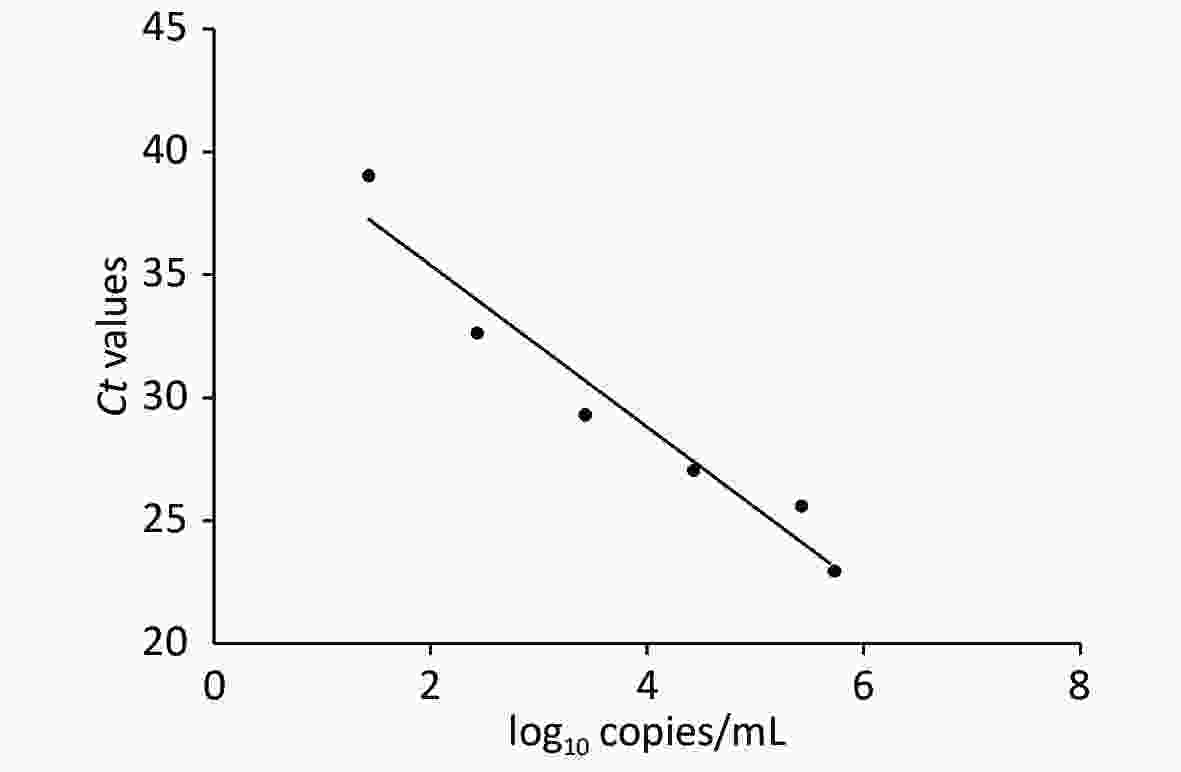
Figure S1. Standard curve of Ct values of real-time RT-PCR against the log10 copies/mL of SARS-CoV-2
Wastewater-based epidemiology (WBE) has been identified previously as a useful tool for disease surveillance with an excellent performance[6]. In the current study, a total of 196 sewage samples were collected from the above mentioned 28 sites (7 samples from each site). During the given period, Lahore had an average temperature of 35 °C (range, 27−41 °C) throughout the day and night. The weather remained sunny overall with a total precipitation of 3.5 mm. The longitude and latitude of the sampling sites of Johar Town C Block and Township A-II sector are provided in Supplementary Table S1, available in www.besjournal.com. The physicochemical parameters of the sewage water samples of the main sites are shown in Table 1. Of the 28 samples, 16 samples were positive on day 1, while 19, 17, 23, 17, 5, and 9 samples were positive on days 3, 5, 7, 9, 11, and 13, respectively. All the results were obtained by real-time PCR, which is considered to be the optimum test to identify SARS-CoV-2[7]. We used a commercially available Sansure kit for viral genome detection as the detection efficacy of the SARS-CoV-2 genome by the Sansure kit is better than that of BGI and Bioperfectus[8].
Table S1. Dimensions of sewage pipelines and longitudes and latitudes of the sampling sites
Sampling sites Line diameter (inches) Longitude Latitude Johar town C-Block Site 1 12 74.29545 31.4524723 Site 2 9 74.2977366 31.4557036 Site 3 9 74.2942093 31.4573861 Site 4 9 74.2989921 31.4552673 Bheer pind LS 42 74.2952135 31.4572428 Township A-II sector Site 1 9 74.2928192 31.435475 Site 2 12 74.294026 31.454058 Site 3 36 74.2929095 31.4352528 Site 4 36 74.2929237 31.4354824 Site 5 9 74.3165282 31.4551185 Site 6 9 74.3157992 31.4552004 Site 7 9 74.3175337 31.4546141 Site 8 18 74.3180418 31.4547766 Site 9 18 74.3166218 31.4552856 Site 10 18 74.3175544 31.4549163 Site 11 9 74.3175511 31.4546758 Site 12 9 74.3189623 31.451292 Site 13 9 74.3191111 31.4515717 Site 14 9 74.3145578 31.4526552 Site 15 9 74.3142842 31.4521259 Site 16 9 74.3157992 31.4552004 Site 17 9 74.3165282 31.4551185 Site 18 9 74.3145578 31.4526552 Site 19 9 74.3175337 31.4546141 Township LS 36 74.3128196 31.4515134 Green town DS 36 74.3125476 31.455127 Ameer chowk DS 36 74.289159 31.4343771 Sadiq chowk DS 36 74.3055126 31.4483829 Note. LS: lift station; DS: disposal station. Table 1. Physicochemical characterization of the sewage water from different sites
Parameters Unit Bheer pind LS Sadiq chowk DS Township LS Ameer chowk DS Green town DS pH − 7.38 7.45 7.57 7.49 7.65 BOD mg/L 320 215 185 365 345 COD mg/L 850 600 535 1,100 1,095 TDS mg/L 805 655 765 940 950 NH3 mg/L 25.8 22.7 33.4 16.7 37.6 Chlorine mg/L BDL 0.4 BDL 0.2 BDL Note. BOD: Biological oxygen demand; COD: Chemical oxygen demand; TDS: Total dissolved solids; BDL: Below detectable level; LS: Lifting station, DS: Disposal station. Of the 5 sampling sites of Johar Town C Block, 4 sites were street sewerage lines, and the 5th site was the relevant sewerage lift station. On the first day of sampling (July 13), all street sewerage pipelines (4/4) were found to be positive for SARS-CoV-2, with a variable load of SARS-CoV-2 genome (102.426 to 104.556 copies/mL) (Figure 1). Sampling site 1 with a 12-inch-diameter pipeline remained positive for the SARS-CoV-2 genome throughout the study, with the highest genome copies 103.64 on the 4th sampling on July 19, 2020. The presumptive reason for this finding is a high amount of passage of sewage water through this pipeline. The SARS-CoV-2 genome was not detected from subsequent samples from sites 2−4 throughout the study period, except for 4th and 5th samples from site 3 (Figure 2). The SARS-CoV-2 genome was not detected from the Bheer Pind lift station of Johar Town in the first sampling on July 13, while two subsequent samples on July 15 and July 17 were detected positive for SARS-CoV-2, with a decreased viral load (102.73 and 0.60 copies/mL, respectively). The varied status of the virus presence from negative to positive can be attributed to the changing disease patterns within the area or movement of asymptomatic carriers within the region. Overall, toward the end of the lockdown period on July 23, SARS-CoV-2 was not detected from 80% (4/5) sewerage sites of Johar Town C Block (Figure 1).
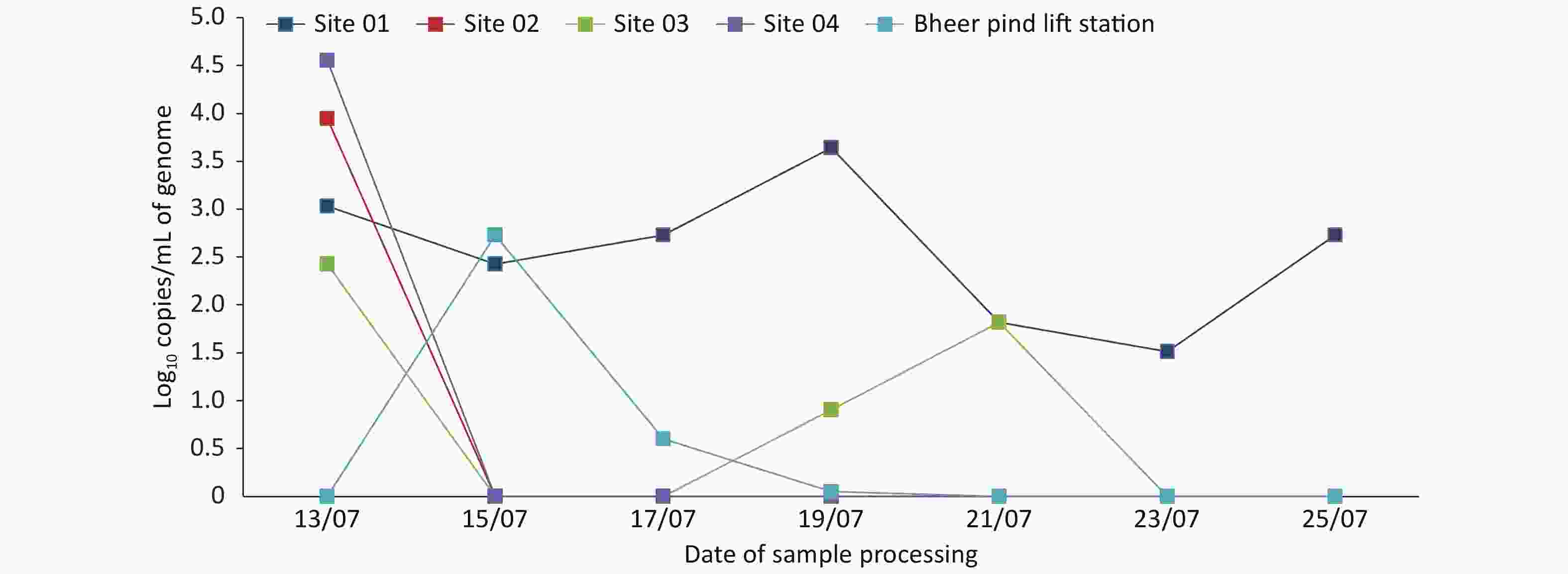
Figure 2. Graphical display of the SARS-CoV-2 viral load expressed as log10 copies/mL at the sites of Johar Town C Block with respect to the progression of lockdown days.
A total of 23 sewerage sites were sampled from Township A-II sector, Lahore, based on the population density and sewerage maps of the area. Among these sites, 19 sites were street sewerage pipelines, while the remaining 4 sites were relevant downstream lift/disposal stations. Of the 19 sites, 10 sites were detected positive for SARS-CoV-2 on the first sampling on July 13, while 16, 11, 16, 11, 4, and 5 sites were identified on the subsequent samplings on July 15, 17, 19, 21, 23, and 25, respectively (Supplementary Table S1, Figure 3). Among the 4 lift/disposal stations of the Township area, 2 stations were detected positive on the first sampling, while all 4 lift/disposal stations were detected negative for SARS-CoV-2 in later samplings (Figure 3). The decrease in viral load in the sewage samples with the progression of smart lockdown was evident by the decreased number of COVID-19 cases reported from the lockdown areas. Before smart lockdown, 1,029 COVID-19 cases were reported from smart lockdown and adjoining areas (Iqbal Town) in the duration of 14 days, while the new cases were decreased to 339 during the smart lockdown.
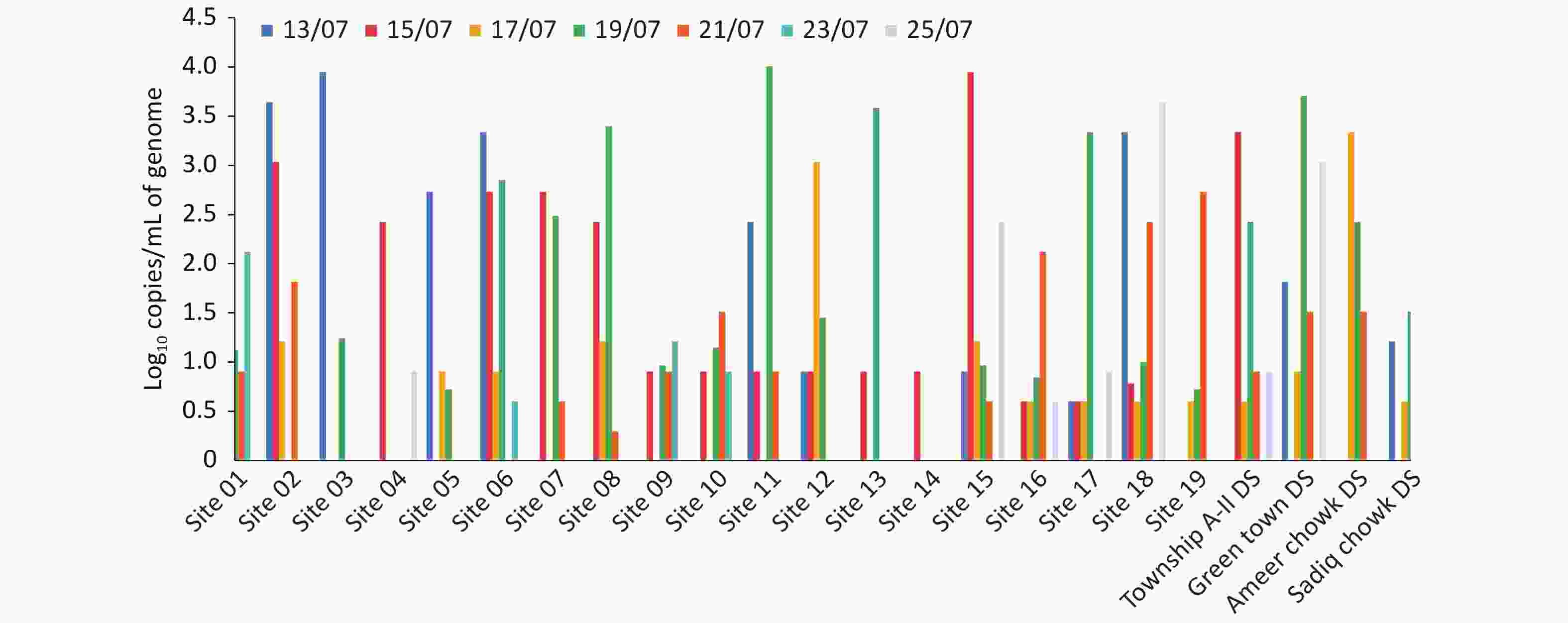
Figure 3. Graphical display of the SARS-CoV-2 viral load expressed as log10 copies/mL at the sites of Township A-II sector with respect to the progression of lockdown days.
To assess the role of sampling time in the detection of SARS-CoV-2 in sewage, two sewerage lift stations, one from each area under smart lockdown, were sampled on an hourly basis for 24 h, and the detection trend of SARS-CoV-2 from hourly samples is presented in Supplementary Figure S2, available in www.besjournal.com. Different sampling time was revealed to be better for both areas. Our results indicate that the best time of sampling may vary between different localities depending on various factors. The results showed that in both regions, more viral genome copies were detected in the early morning (7 am to 10 am) and evening to midnight (6 pm to 12 am). This may be due to the peak in toilet use in the morning and evening[9]. These results suggest that an appropriate sampling time should be selected by using the hourly sampling method.
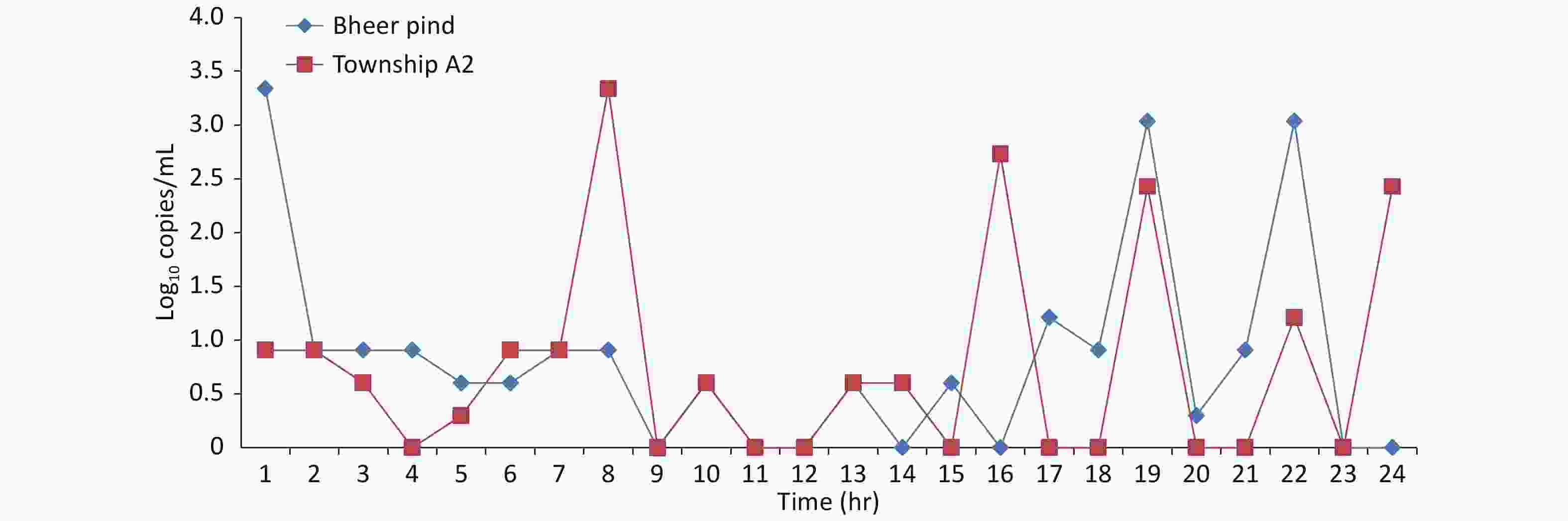
Figure S2. Graphical display of the SARS-CoV-2 viral load as log10 copies/mL at Site 05 of Johar Town and Site 20 of Township with respect to time in hours starting from 1:00 am for the period of 24 hours.
The constant positive status of site 1 of Johar Town C Block throughout the lockdown period prompted the backtracking of the upstream sewage water, which led us to the homes with active cases of COVID-19. The sewage of 16/67 homes was found to be positive for SARS-CoV-2. Residents of 5 homes were found to be symptomatic and positive for nasopharyngeal or serum detection, while 127 asymptomatic residents of the 67 homes were positive for either nasopharyngeal swab or antibodies against the virus. SARS-CoV-2 has been previously detected in fecal samples of both symptomatic and asymptomatic patients of COVID-19[10].
The success of identifying the homes containing active patients of COVID-19 by backtracking makes our approach to be valid for optimizing lockdowns. One of the striking observations of the study is that SARS-CoV-2 RNA can be detected in raw sewage water samples, without the need to concentrate the virus. Collectively, this study indicates that sewage water-based COVID-19 surveillance could play a reliable role in the monitoring and execution of smart lockdowns.
We acknowledge the WASA Directorate, Lahore region for facilitating sampling across the district. We thank the Primary and Secondary Healthcare Department, Punjab for the financial support to this project
The authors have no conflicts of interest to disclose.
全文HTML
 21030.pdf
21030.pdf
|

|



 下载:
下载:



 Quick Links
Quick Links