-
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a severe global pandemic. Based on the data from the World Health Organization (WHO), as of May 19, 2024, more than 775 million confirmed cases of COVID-19 registered globally, with more than 7 million deaths[1].
SARS-CoV-2 is a positive-sense single-stranded RNA virus with a 30 kb genome, and it is highly susceptible to sequence mutations caused by random errors during genetic replication. Currently, with the accumulation of viral mutations, SARS-CoV-2 has evolved into divergent variants including Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2) and Omicron (B.1.1.529). Omicron with the increasing infectivity and transmissibility has displaced Delta and become predominant variant worldwide. Subsequently, the sub-lineage BA.2 has spread to many countries and regions worldwide and become dominant variant in Western countries[2]. Compared with the Wuhan-Hu-1 reference genome, the RBD of BA.2 variant contained 16 mutations, including G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, and Y505H, which were associated with the high infectivity and immune resistance of vaccines[2,3]. Furthermore, most of the prevalent subvariants including BA.4/5, XBB, JN.1, and KP.2 belong to the phylogenetic clade related to BA.2 variant, which exhibited immune escape from vaccines and monoclonal antibodies (mAbs)[3-5]. More importantly, Muik and his colleagues found that BA.2 breakthrough infection enhanced the cross-neutralization of BA.2.12.1 and BA.4/5 sublineages[6]. Therefore, the development of BA.2 effective vaccines would be conducive to provide scientific basis for the prevention the infection of BA.2 descendants and recombinant subvariants.
Presently, 183 vaccines have entered into the clinical studies, including 59 protein subunit vaccines, 25 non-replicating viral vector vaccines, 22 inactivated vaccines, and 60 nucleic acid vaccines (RNA and DNA)[7]. Compared with inactivated vaccines, recombinant subunit vaccines were of safety profile and induced the highly specific immune responses, which played a crucial role in preventing COVID-19 in individuals as soon as the epidemic began[8]. Spike (S) protein especially RBD was crucial for mediating viral attachment, entry and virus-host membrane fusion by specifically binding to human angiotensin-converting enzyme 2 (hACE2) on the surface of host cells[9]. The RBD that contained abundant T- and B-cell epitopes induced the NAbs and cellular immune responses, which was the primary target for the development of subunit vaccines.
A universal P2 T-helper cell epitope (TT-P2), with activity comparable to intact Tetanus toxoid (TT), was of promiscuous binding to several different human leukocyte antigen (HLA) DR molecules, which has been widely used as an immunostimulant to enhance the cellular immune responses of subunit vaccines[10,11]. Therefore, TT-P2 was used as an endogenous adjuvant to improve the immune responses induced by RBD recombinant protein. In addition, CpG 1826 as a novel adjuvant have been used widely to enhance the immune responses of influenza, HIV and malaria vaccines[12-14]. Meanwhile, several studies have revealed that CpG plus alum adjuvant could synergistically strengthen the immune responses of vaccines and balance the Th1/Th2 immune responses[15-18]. In this study, to obtain the required a large of immunogens through an accessible, rapid and cost-efficient approach, Escherichia coli (E. coli) expression system was used to prepare the recombinant protein designated as Sot, which contained the TT-P2 epitope linked to the carboxy-terminal of the BA.2 variant monomeric RBD (residues 308-548) through flexible linker. Then the immunogenicity of soluble Sot protein formulated with CpG 1826 plus alum dual adjuvant (designated as Sot/CA) was evaluated in BALB/c mice.
-
All animal experimental procedures in this study were approved by the Animal Care and Welfare Committee of the National Institute for Viral Disease Control and Prevention at the Chinese Center for Disease Control and Prevention (Approval No. 20230609046).
-
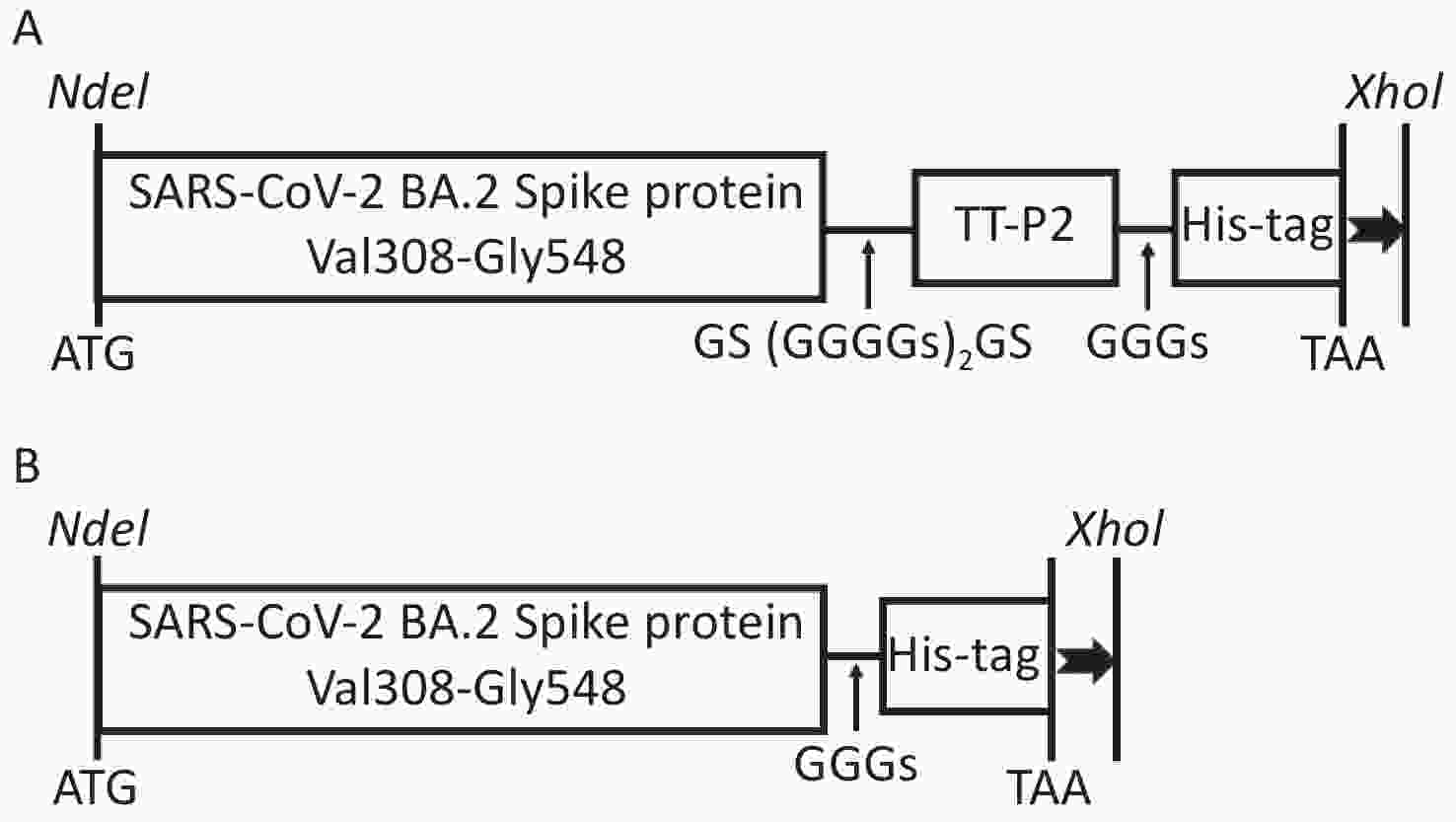
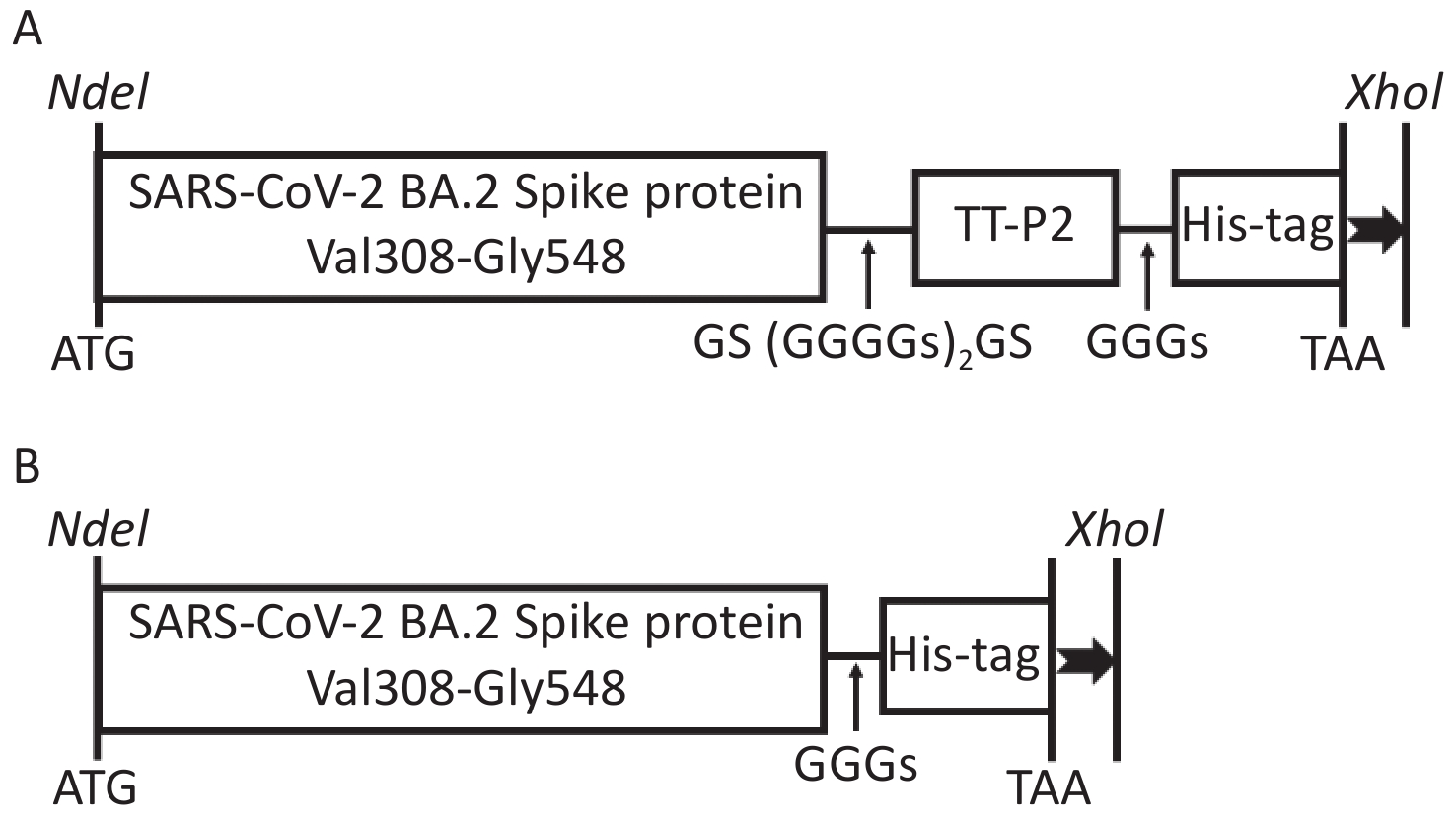
The S glycoprotein amino acid sequence of the SARS-CoV-2 Omicron BA.2 variant (GenBank Accession No. UKW53095.1) was obtained from the National Center for Biotechnology Information (NCBI) database. The recombinant protein comprising the Val308-Gly548 fragment of S glycoprotein attached to the TT-P2 epitope with the flexible linker GS(GGGGS)2GS was designated as Sot (Figure 1A), while the recombinant protein containing only the Val308-Gly548 fragment was designated as So (Figure 1B). A 6 × His tag was fused to the C-terminal of the recombinant protein to facilitate purification. The codon-optimized gene sequences were artificially synthesized (General Biol, Anhui Chuzhou, China) and then subcloned into the pET30a(+) vector by using restriction endonucleases NdeI and XhoI (New England Biolabs). The positive recombinant plasmids identified by agarose gel electrophoresis and DNA sequencing were transformed into E. coli BL21 (DE3) competent cells.
-
Fresh transformed E. coli BL21 (DE3) cells containing the recombinant plasmids were grown in 3 mL Luria-Bertani (LB) medium for 6 h and then transferred into 3 L LB medium with 1:1,000 for 3 h. The expression was induced by adding 1 mmol/L isopropyl β-D-thiogalactopyranoside (IPTG) at 37 °C for 3 h. Protein expression was confirmed by the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The cells were harvested by centrifugation (8,000 ×g, 10 min, 4 °C). The cell pellets were resuspended in 50 mL of Buffer A (20 mmol/L Tris-HCl, 500 mmol/L NaCl, 10% glycerol, 1% NP-40, pH 8.0), followed by ultrasonic disruption (300 W, 20 s, 20 times) (Scientz, Zhejiang Ningbo, China). Following centrifugation (8,000 ×g, 10 min, 4 °C), the pellets were dissolved in Buffer B (20 mmol/L Tris-HCl, and 8 mol/L urea, pH 8.0). The denatured inclusion bodies (IBs) were purified by DEAE ion exchange chromatography (IEC) and Ni-NTA affinity chromatography (GE Healthcare, USA), respectively. The IBs was dialyzed in Buffer C (20 mmol/L Tris-HCl, 10% glycerol, 500 mmol/L NaCl, 0.1 mmol/L EDTA, 0.3 mol/L arginine, 0.4 mmol/L oxidized glutathione, 4 mmol/L reduced glutathione, pH 8.0) to gradually decrease the urea content. The purity and concentration of the renatured protein were determined by High Performance Liquid Chromatograph (HPLC) and bicinchoninic acid (BCA) kit (TransGen Biotech, Beijing, China), respectively. According to the manufacturer’s instructions, the endotoxin of the proteins was removed by the commercial ToxinEraser™ Endotoxin Removal Kit (GenScript, USA) and then the ToxinSensor™ Chromogenic Limulus Amebocyte Lysate (LAL) Endotoxin Assay Kit (GenScript) was used to quantify the concentration of Lipopolysaccharide (LPS) of the proteins[19].
-
Soluble proteins were loaded on 12% SDS-PAGE and then transferred to the polyvinylidene difluoride membrane. The membrane was probed with the commercial anti-SARS-CoV-2 RBD antibodies (Abcam, England, ab277628) (1,000-fold diluted in 5% skimmed milk). The horseradish peroxidase (HRP)-conjugated goat anti-mouse antibodies (2000-fold diluted) (Beyotime, Shanghai, China) was used as the secondary antibody[20].
-
The binding of proteins to serum IgG antibodies was evaluated by ELISA. Briefly, the 96-microwell plates were coated with the soluble proteins (0.2 μg/well) at 4 °C overnight. The plates were washed five times with phosphate-buffered saline containing 0.05% Tween-20 (PBST), pH 7.4 and then blocked with 5% skimmed milk in PBST at 37 °C for 2 h. Washed by PBST, 100 μL of 100-fold diluted serum from healthy donors (HDs) or COVID-19 patients was added and incubated at 37 °C for 2 h. Washed by PBST again, the HPR-conjugated anti-human IgG antibodies (5000-fold diluted) was added and incubated at 37 °C for 1 h. Washed again and then 3,3’,5,5’,-tetramethylbenzidine (TMB) substrate (Solarbio, Beijing, China) was added and incubated for 15 min in the dark. The reaction was terminated with the 100 μL of 2 mol/L H2SO4. Finally, the absorbance at 450 nm was measured by using an ELISA reader (Thermo Fisher Scientific, USA)[20].
-
CpG 1826 (5’-TCCATGACGTTCCTGACGTT-3’) was synthesized by SYNBIO Technologies (Suzhou, China) and completely dissolved in 20 mmol/L Tris-HCl (pH 8.0). The CpG 1826 plus alum was designated as CA dual adjuvant with the final concentration of 0.5 mg/mL, respectively. For the immunogenicity evaluation, the 40 μg of So or Sot protein was thoroughly emulsified with 50 μg of CpG 1826, alum or CA dual adjuvant, respectively.
Specific-pathogen-free (SPF), female BALB/c mice (aged 6–8 weeks) were purchased from Vital River Laboratories (Beijing, China) and randomly divided into 9 groups (n = 5/group). Mice were immunized intramuscularly with 100 μL of So/alum, Sot/alum, So/CpG 1826, Sot/CpG 1826, So/CA, or Sot/CA on day 0 and 14, while negative controls were immunized with an equal volume of the CA dual adjuvant, CpG 1826 or alum adjuvant without the immunogens. Mice were then sacrificed for the immunogenicity evaluation on day 28 after the prime vaccination. The animal grouping and prime-boost vaccination regimen are shown in Table 1.
Table 1. Animal grouping and vaccination regimen of mice
Group Adjuvant Route Dose Interval N# Adjuvant CpG 1826+alum i.m. 100 μL/50 μg 0, 14 5 Adjuvant CpG 1826 i.m. 100 μL/50 μg 0, 14 5 Adjuvant alum i.m. 100 μL/50 μg 0, 14 5 So/alum alum i.m. 40 μg/50 μg 0, 14 5 Sot/alum alum i.m. 40 μg/50 μg 0, 14 5 So/CpG 1826 CpG 1826 i.m. 40 μg/50 μg 0, 14 5 Sot/CpG 1826 CpG 1826 i.m. 40 μg/50 μg 0, 14 5 So/CA CpG 1826+alum i.m. 40 μg/50 μg 0, 14 5 Sot/CA CpG 1826+alum i.m. 40 μg/50 μg 0, 14 5 Note. i.m.: intramuscular injection; N#, number of mice. -
The specific anti-RBD antibody responses of immunized mice were evaluated by indirect ELISA. Orbital blood samples were collected from mice and naturally coagulated at room temperature (RT) for 4 h. Serum was then obtained from the coagulated blood samples by centrifugation at 4,000 ×g for 10 min. Next, plates were coated with the Sot or So protein (100 ng/well) at 4 °C overnight. The plates were washed by PBST and then blocked with 5% skimmed milk in PBST at 37 °C for 2 h. Washed by PBST again, serially diluted mice serum (from 24 to 220) was added and the plates were incubated at 37 °C for 2 h. Washed again, the HRP-conjugated goat anti-mouse IgG, IgG1, and IgG2a antibodies (diluted 5000-fold in 5% skimmed milk) were added with incubating at 37 °C for 1 h. Washed again, TMB substrate was added and the color reaction was terminated by adding 2 mol/L H2SO4. Finally, the absorbance at 450 nm was measured after 30 min. The highest dilution at which the mean absorbance of the sample was 2.1-fold greater than that of the negative control serum was considered the antibody titer[21].
-
ELISpot assay was conducted to determine the cellular immunity in immunized mice by quantifying IFN-γ and IL-4-specific splenocytes according to the manufacturer’s instructions (MabTech, Sweden). Briefly, the splenic lymphocytes of immunized mice, separated by a commercial kit (TBD Science, Tianjin, China) through density gradient centrifugation, were seeded into 96-well plates (2 × 105 cells/well, in triplicates) and stimulated by an equivalent volume of concanavalin A (positive control, 2 μg/well), the So/Sot protein (3 μg/well) or RPMI 1640 medium (negative control), respectively. After incubation for 20 h at 37 °C in a humidified incubator with 5% CO2, the cells were removed and washed with PBS (200 μL/well). Subsequently, biotin-conjugated detection antibodies (1:1,000 in PBS containing 0.5% fetal calf serum) were added and the plates were incubated at RT for 2 h. Washed by PBS, streptavidin-ALP (1:1,000 diluted) was added and the plates were incubated at RT for 1 h. Next, the substrate solution was filtered and then added (100 μL/well). After distinct spots emerged on the plates, color development was stopped by washing extensively with deionized water. Finally, the plates were left to dry and the numbers of spot-forming cells (SFCs) were counted by an ELISpot reader (AID, Germany).
-
The geometric mean titer (GMT) of NAbs was detected as previous study[21]. In brief, 50% tissue-culture infectious dose (TCID50) units of BA.2 and XBB.1.5 virus were mixed with an equal volume of 2-fold serial diluted (from 22 to 212) mice sera and incubated at 37 °C for 2 h, respectively. The virus-serum mixture was added to the monolayers of Vero E6 cells (1 × 105 cells/well) and then incubated for 72 h. Cytopathic effect (CPE) was recorded using an inverted microscope. The GMT of NAbs was calculated as the highest serum dilution that completely prevented CPE in 50% of the wells according to the Reed-Meunch method[22].
-
The experimental data were analyzed by GraphPad Prism, version 8.0 (GraphPad Software, California, USA). The GMT was used to represent the antibodies titer. The numbers of SFCs and the GMT of antibodies between the different groups were compared by Two-way ANOVA and Tukey’s multiple comparison test. P ≤ 0.05 was considered to determine statistically significant difference.
-
The pET30a(+)-So and pET30a(+)-Sot recombinant plasmids containing So gene fragment (774 bp) and Sot gene fragment (858 bp) were identified by 1% agarose gel electrophoresis, respectively. This result consistent with DNA sequencing indicated that the So and Sot gene fragment were correctly subcloned into pET30a(+) vector (5,234 bp), respectively (Figure 2).
Figure 2. Agarose gel electrophoresis of the recombinant plasmids: Lane M, DNA Marker; lane 1, pET30a(+)-So recombinant plasmid; lane 2, pET30a(+)-Sot recombinant plasmid.
The His-tagged recombinant proteins were expressed as the form of IBs. The theoretical molecular weight (MW) of the So and Sot proteins were 28 and 31 kD, respectively, which were consistent with the SDS-PAGE analysis (Figures 3A and 3B). The purity of So and Sot proteins were more than 95% with the concentrations of 0.54 and 0.50 mg/mL. According to the LAL method, the endotoxin concentration of proteins was less than 1 EU/mL.
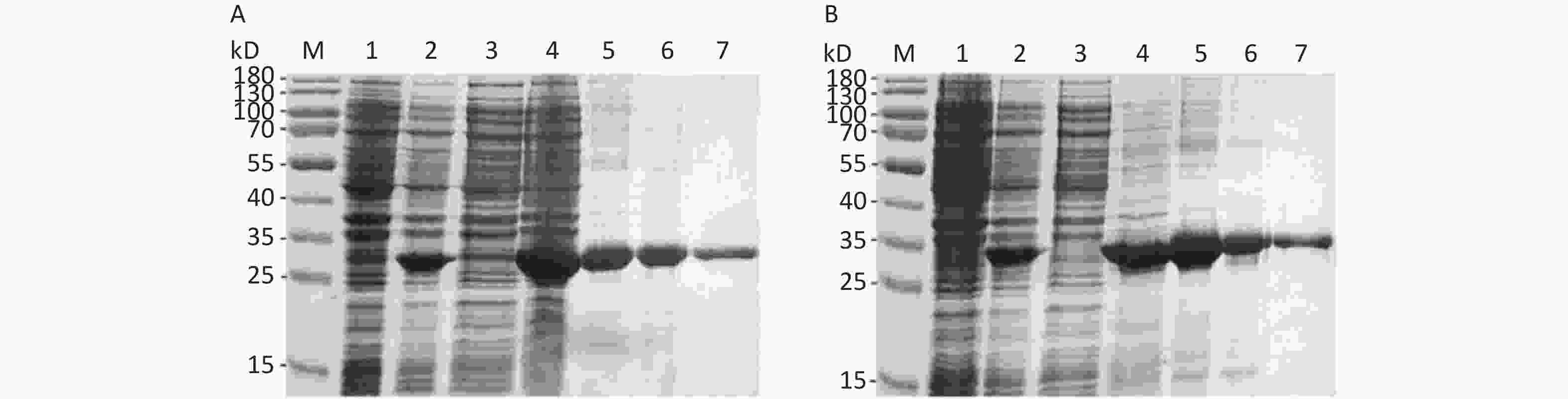
Figure 3. Expression and purification of recombinant proteins. (A) SDS-PAGE analysis of the recombinant So protein: Lane M, protein marker; lane 1, non-induced bacteria; lane 2, induced bacteria; lane 3, soluble fraction; lane 4, IBs; lane 5, denatured So protein; lane 6, So protein purified by IEC; lane 7, So protein purified by Ni-NTA. (B) SDS-PAGE analysis of the recombinant Sot protein: Lane M, protein marker; lane 1, non-induced bacteria; lane 2, induced bacteria; lane 3, soluble fraction; lane 4, IBs; lane 5, denatured Sot protein; lane 6, Sot protein purified by IEC; lane 7, Sot protein purified by Ni-NTA. SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; IBs, inclusion bodies; IEC, ion-exchange chromatography.
-
Western Blotting (WB) assay showed that two signal bands of the So and Sot protein at 28 and 31 kD, respectively, suggesting that the recombinant proteins could be specifically recognized by the anti-RBD mAbs (Figure 4A). In addition, the antigenicity analysis of So and Sot recombinant proteins were assessed by ELISA, respectively. So and Sot proteins showed that similarly specific reactivity with COVID-19 positive serum, suggesting that the recombinant proteins possessed the antigenic properties (Figure 4B).
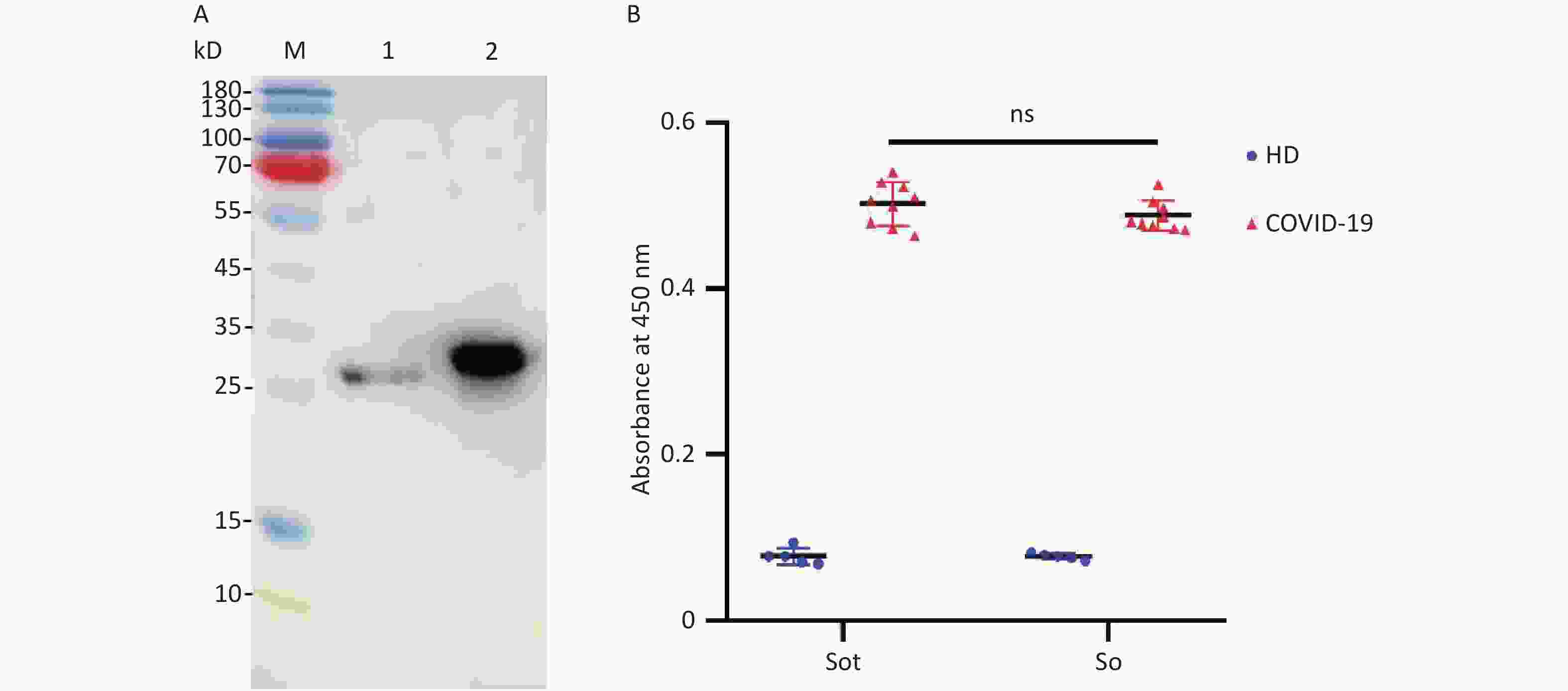
Figure 4. Antigenicity analysis of the recombinant proteins: (A) WB assay of the proteins: Lane M, protein marker; lane 1, So protein; lane 2, Sot protein. (B) Absorbance at 450 nm of ELISA. WB, Western blotting; ELISA, enzyme-linked immunosorbent assay; HD, healthy donors; COVID-19, coronavirus disease 2019.
-
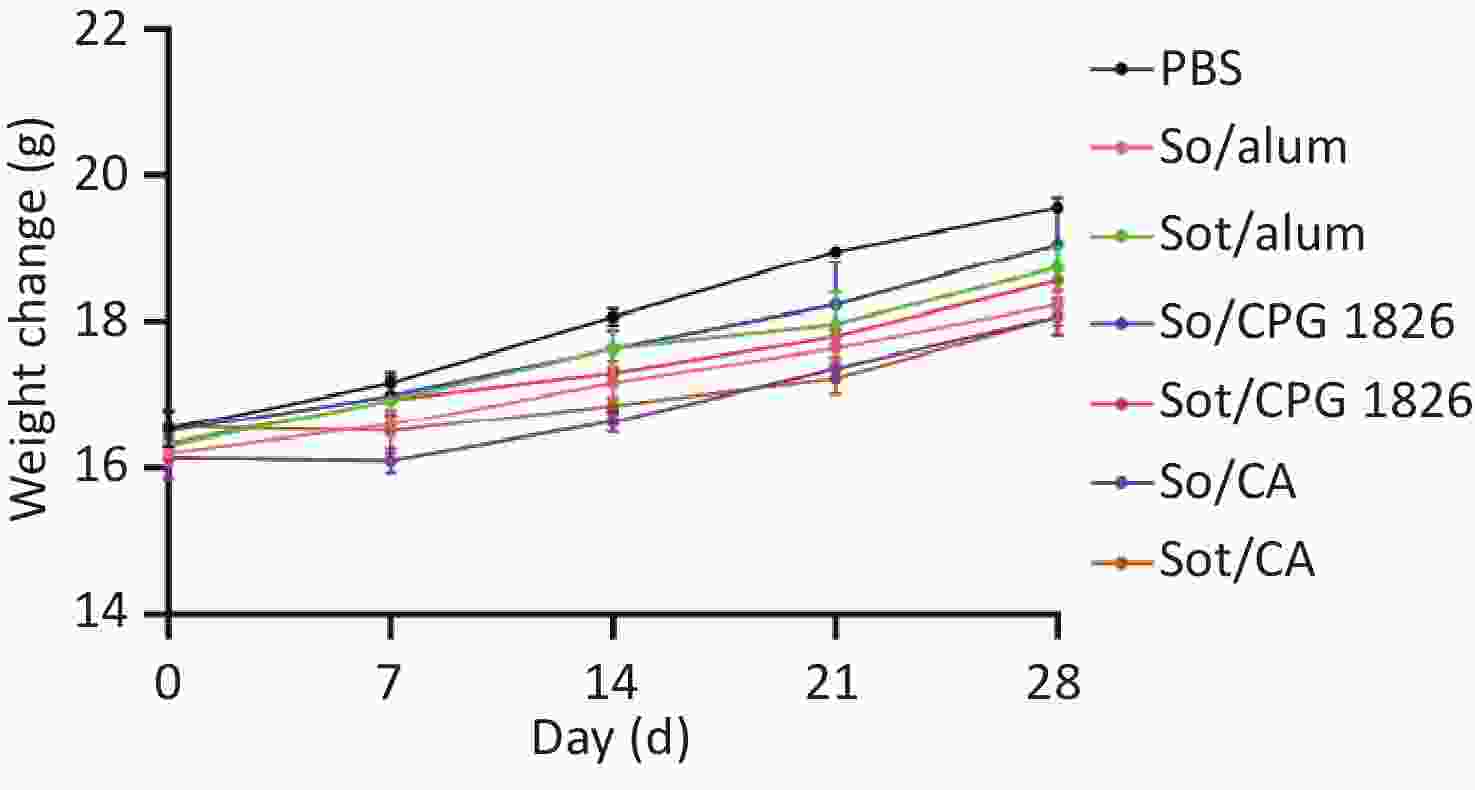
There was no mortality and the body weight of immunized mice gradually increased during the immunization. Although the body weight of Sot/CA and So/CA groups decreased on day 7, the mean weight of mice was no significant difference compared with the other groups (P > 0.05). Moreover, the water and food intake of immunized mice of each group was no obvious abnormal, indicating that subunit vaccines had no adverse effect on the survival and viability of immunized mice during the immunization period (Figure 5).
-
IgG antibodies were detected by indirect ELISA to evaluate humoral immunogenicity in immunized mice. As shown in Figure 6, no antigen-specific IgG antibodies were detected in the negative adjuvant groups, while in the subunit vaccines groups, high titers of IgG antibodies were detected and drastically increased with the booster vaccination. On day 28, the GMT of IgG antibodies in the Sot/CA group (75,281) was 4- and 16-fold higher than that in the Sot/CpG 1826 (18,820) and Sot/alum group (4,705), respectively. Similarly, the GMT of IgG antibodies in the So/CA group (24,833) was 8- and 14-fold higher than that in the So/CpG 1826 (3,104) and So/alum group (1,783), respectively. These results showed that the CA dual adjuvant synergistically enhanced the IgG antibody responses induced by the subunit vaccines. In addition, in the CA dual adjuvant groups, the GMT of IgG antibodies induced by Sot protein was 3-fold higher than that induced by So protein (P < 0.01). Similarly, whether in the alone CpG 1826 or alum adjuvant group, the GMT of IgG antibodies induced by Sot protein was 6- and 2.6-fold higher than that by So protein, respectively.
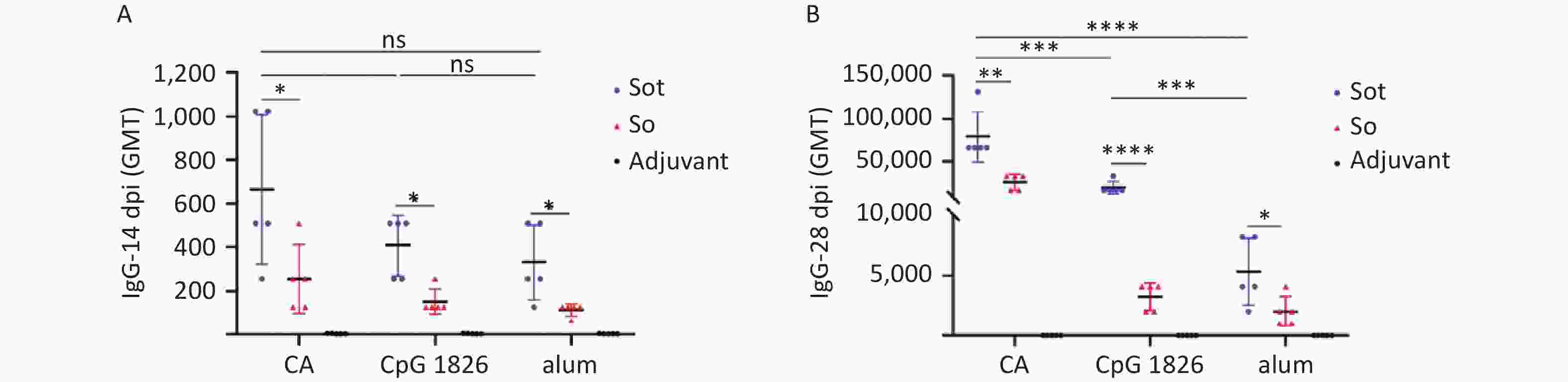
Figure 6. The titer of specific IgG antibodies in immunized mice. (A) The titer of IgG antibodies on day 14. (B) The titer of IgG antibodies on day 28. dpi, day post immunization; GMT, geometric mean titer. The data were analyzed to evaluate significant differences by Two-way ANOVA. Asterisk (*) represents the difference between the experimental groups. ns (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
-
IgG antibody isotypes were also detected by indirect ELISA. As shown in Figure 7A, subunit vaccines formulated with CA dual adjuvant induced higher titers of IgG1 antibodies than those with alone CpG 1826 or alum adjuvant (P < 0.0001). Meanwhile, regardless of the adjuvant, the titer of IgG1 antibodies induced by Sot protein was slightly higher than that induced by So protein (P < 0.05). As shown in Figure 7B, subunit vaccines formulated with CA dual adjuvant induced higher titers of IgG2a antibodies than those with CpG 1826 or alum adjuvant (P < 0.0001). Meanwhile, in the CA dual adjuvant and CpG 1826 adjuvant groups, the GMT of IgG2a antibodies induced by Sot protein was higher than that induced by So protein (P < 0.05), however, there was no significant difference of IgG2a antibodies between Sot and So protein in the alum adjuvant group (P > 0.05) (Figure 7B).
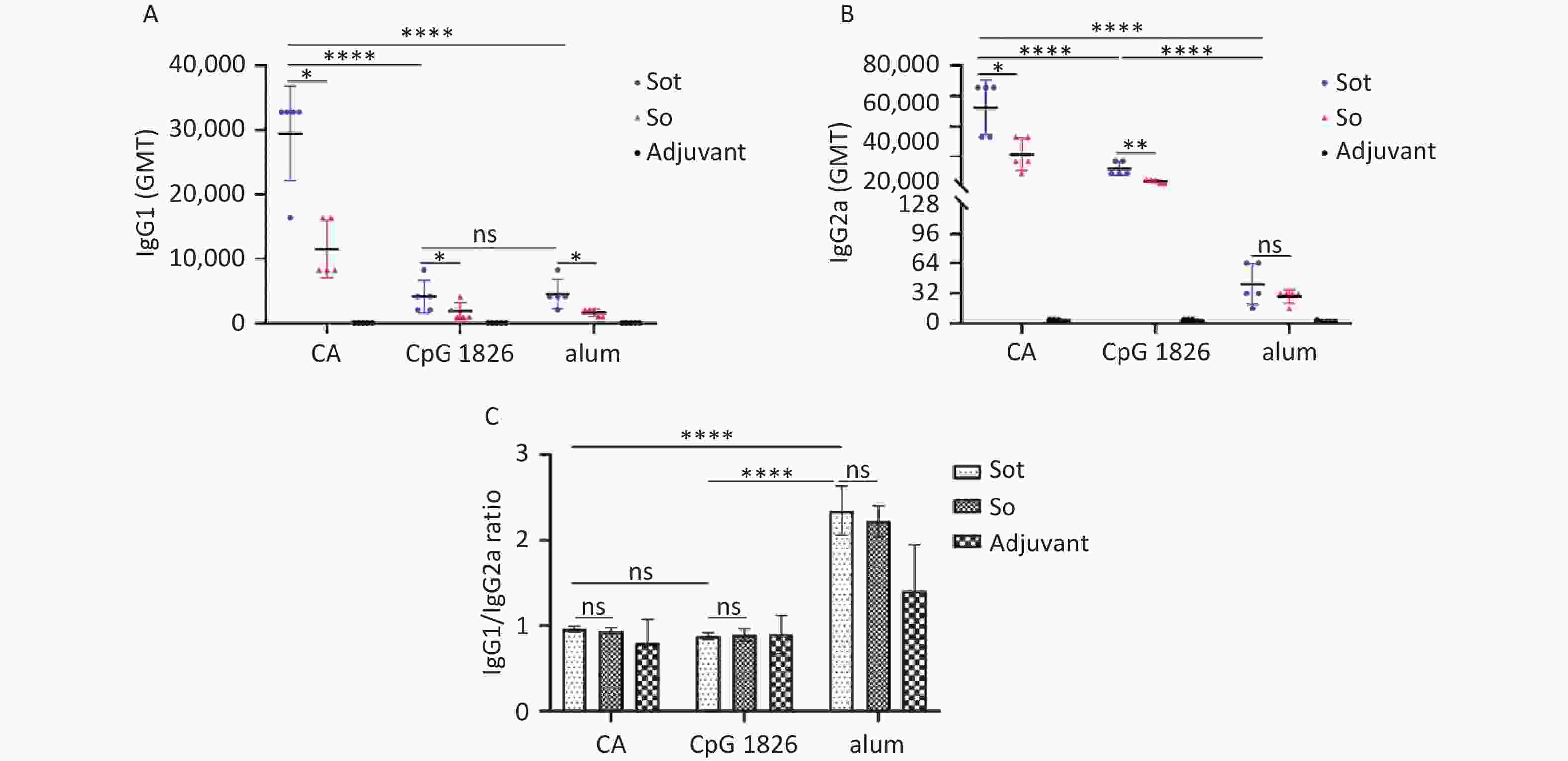
Figure 7. Analysis of IgG antibody isotypes in immunized mice. (A) The titer of IgG1 antibodies. (B) The titer of IgG2a antibodies. (C) The IgG1/IgG2a ratios in mice serum. GMT, geometric mean titer. The data were analyzed to evaluate significant differences by Two-way ANOVA. Asterisk (*) represents the difference between the experimental groups. ns, P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001.
IgG1 is the main antibody isotype of Th2-type immune response related to the secretion of IL-4, while the IgG2a antibody is associated with Th1-type immune response with secretion of IFN-γ. Therefore, the IgG1/IgG2a ratio was also calculated to assess the type of immune response induced by the subunit vaccines in mice (Figure 7C). The IgG1/IgG2a ratio of the Sot/alum group was 2.34 ± 0.28, which was considerably higher than that of the Sot/CA (0.96 ± 0.03) or Sot/CpG 1826 (0.88 ± 0.04) groups, respectively (P < 0.0001). Similarly, the IgG1/IgG2a ratio of the So/alum group was 2.22 ± 0.18, which was considerably higher than that of the So/CA (0.94 ± 0.03) or So/CpG 1826 (0.9 ± 0.07) groups, respectively (P < 0.0001). However, regardless of the adjuvant, no significant difference in the IgG1/IgG2a ratio was observed between the Sot and So proteins (P > 0.05).
-
The microneutralization assay was conducted to determine the GMT of NAbs against BA.2 and XBB.1.5 variants induced by subunit vaccines. The GMTs of NAbs induced by the Sot/CA and So/CA groups against the BA.2 variant were as high as 1,176 and 676, which were higher than those formulated with alone CpG 1826 or alum adjuvant, respectively. In addition, regardless of the adjuvant, the NAbs induced by Sot protein were significantly higher than those induced by So protein (P < 0.05) (Figure 8A).
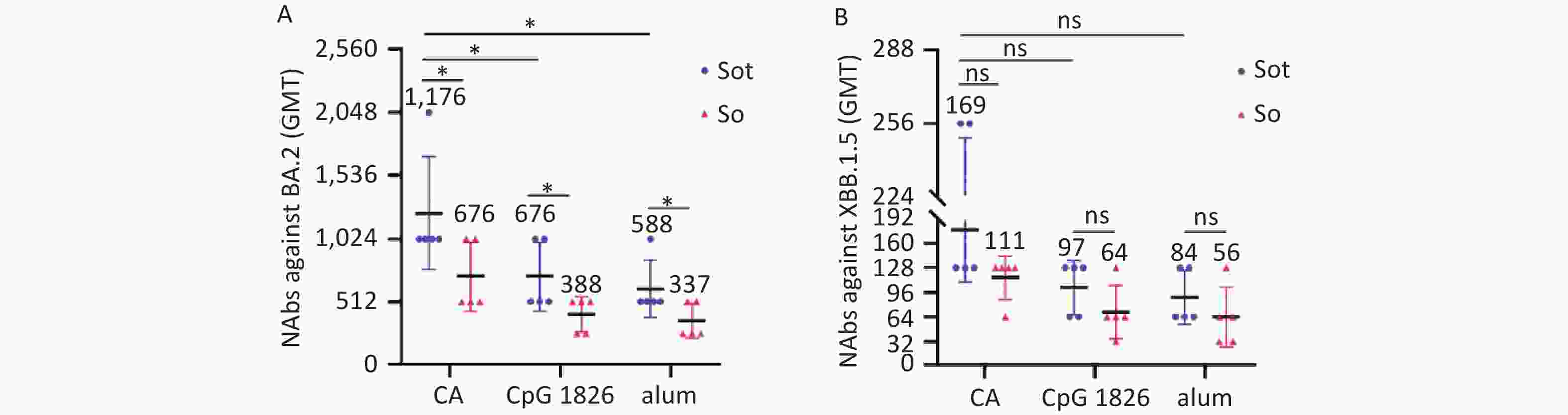
Figure 8. The titer of NAbs in mice serum. (A) The titer of NAbs against BA.2 variant. (B) The titer of NAbs against XBB.1.5 variant. NAbs, neutralizing antibodies; GMT, geometric mean titer. The data were analyzed by Two-way ANOVA to evaluate significant differences. Asterisk (*) represents the difference between the experimental groups. ns, no significance; P > 0.05; *P < 0.05.
Comparatively, the GMTs of NAbs induced by subunit vaccines against the XBB.1.5 variant were significantly lower than those against the BA.2 variant (Figure 8B). The GMTs of NAbs induced by the Sot/CA and So/CA groups against XBB.1.5 variant were 169 and 111, respectively. However, regardless of the adjuvant, there was no significant difference between the subunit vaccines (P > 0.05).
-
To evaluate the cellular immunity, we conducted ELISpot assay to quantify the effector T lymphocytes secreting IFN-γ or IL-4 in mice, respectively. These results showed that Sot protein induced considerably stronger cellular immune response than So protein in immunized mice, regardless of the adjuvant (P < 0.0001). The numbers of IFN-γ SFCs in the Sot/CA and So/CA groups were 635.00 ± 17.62 and 452.20 ± 26.90, respectively, which were significantly higher than those in the alone CpG 1826 adjuvant group (382.40 ± 16.10 and 255.00 ± 14.20) or alum adjuvant group (213.70 ± 0.58 and 94.33 ± 16.77), respectively (Figure 9A) (P < 0.0001). In contrast, the numbers of IL-4 SFCs in the Sot/alum and So/alum groups were 311.7 ± 1.53 and 185.7 ± 4.16, which were significantly higher than those in the CA dual adjuvant group (279.20 ± 13.10 and 153.8 ± 7.86) or CpG 1826 adjuvant group (180.60 ± 4.78 and 103.40 ± 2.30), respectively (Figure 9B) (P < 0.0001).
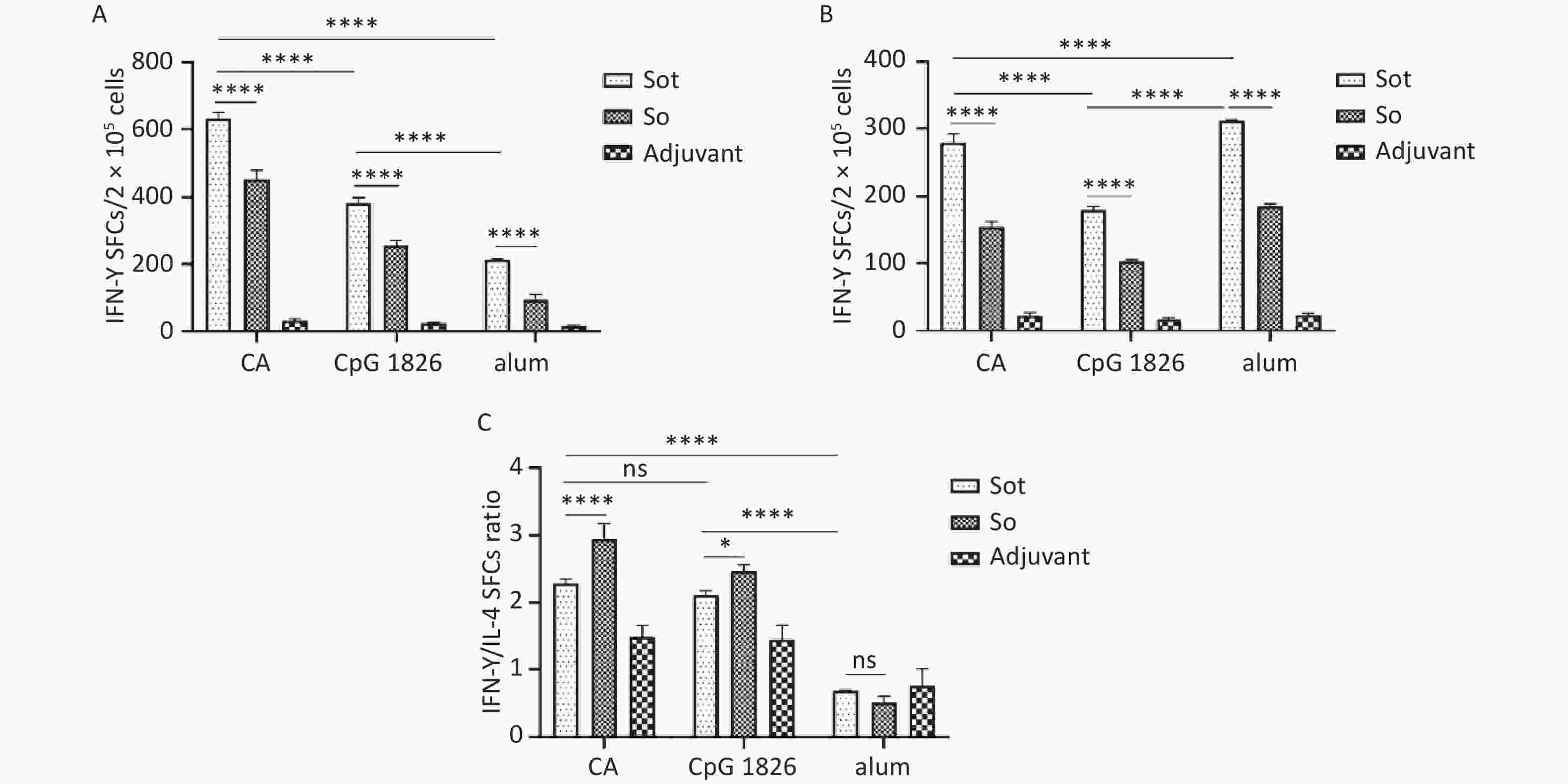
Figure 9. The SFCs secreting IFN-γ and IL-4 in mice spleen. (A) The SFCs of IFN-γ. (B) The SFCs of IL-4. (C) The ratios of IFN-γ/IL-4 SFCs. SFCs, spot-forming cells. The data were analyzed by Two-way ANOVA to evaluate significant differences and expressed as mean ± SD. Asterisk (*) represents the difference between the experimental groups. ns, no significance; P > 0.05, *P < 0.05, ****P < 0.0001.
Additionally, the ratios of IFN-γ SFCs/IL-4 SFCs in the Sot/CA and So/CA groups were 2.28 ± 0.07 and 2.95 ± 0.23, respectively, which were higher than those in the alone CpG 1826 adjuvant group (2.11 ± 0.07 and 2.46 ± 0.09) or alum adjuvant group (0.69 and 0.51 ± 0.10), respectively (Figure 9C).
-
The ongoing COVID-19 pandemic posed a frightening threaten to the health of global citizens. Although the epidemic trend of BA.2 variant has stabilized, a variety of subvariants such as BA.4/5, XBB and JN.1 belong to BA.2 clade have further exacerbated the burden of global COVID-19 prevention and control[3,23]. Consequently, the development of BA.2 vaccines provided a scientific basis for the prevention of COVID-19 caused by BA.2 subvariants.
The prokaryotic expression system was the most widely used recombinant proteins production system, which represented by E. coli has irreplaceable advantages over other expression systems, including the clear genetic background, simple culture conditions and high yield. According to the WHO, several SARS-CoV-2 subunit vaccine candidates based on the E. coli expression system have entered the pre-clinical studies[7]. Nowadays, some vaccines based on the E. coli expression system have been used in human vaccination as authorized by the National Medical Products Administration China[24,25]. Although mRNA vaccines were more prevailing among these platforms of developing SARS-CoV-2 vaccines, considering the cost and accessibility of vaccines, the prokaryotic expression system was a promising platform to develop subunit vaccines, especially in the low- and middle-income countries, which enabled to rapidly prepare a large of immunogens, especially in the case of the various SARS-CoV-2 variant infections.
In this study, recombinant proteins were successfully prepared by E. coli expression system. Since the high level of expression in E. coli cells and the presence of four disulfide bonds between residues Cys336-Cys361, Cys379-Cys432, Cys391-Cys525, and Cys480-Cys488, the recombinant proteins were expressed as the form of IBs, which was consistent with the previous study[21]. Subsequently, the high-purity of soluble proteins were successfully obtained by the chromatography purification technology system and dialysis renaturation. The additives including arginine and redox-shuffling agents such as reduced and oxidized glutathione were conducive to the formation of native disulfide bonds and improving of refolding yield[26,27]. More importantly, soluble proteins with antigenic properties showed that specific reactivity to anti-RBD antibodies and serum of COVID-19 patients (Figure 4).
However, there were some limitations about the prokaryotic expression system. Firstly, the recombinant proteins expressed by E. coli were non-glycosylated. Although glycosylation played an important role in the recognition of host cell receptors, viral assembly and induction of immune responses, the mechanism of interaction between the degree of glycosylated modification and vaccine efficacy remained to be further investigated[28]. There were only two potential O-linked (residues 323 and 325) and two N-linked (residues 331 and 343) glycosylated sites in the Val308-Gly548 fragment, which were distant from binding to hACE2[29]. Furthermore, Huang et al. have demonstrated that non-glycosylated S protein formulated with alum adjuvant could induce the cellular and humoral immune responses, which were superior to those of glycosylated S protein vaccines; meanwhile, immune serum of mice was of great cross-neutralizing activity against Alpha, Gamma and Delta variants[30]. More importantly, some non-glycosylated RBD recombinant proteins showed the great immunogenicity to induce the robust immune responses[21,31]. Therefore, it was reasonable to deduce that the glycosylated modification might not be the most critical factor that impacting the immunogenicity of vaccines. Secondly, the proteins obtained by E. coli were susceptible to LPS contamination, and the additional manipulations were required to purify the protein and eliminate endotoxins.
Antibody-mediated humoral immune responses played a crucial role in defensing SARS-CoV-2 infection. ELISA results showed that Sot/CA subunit vaccine had the advantage of inducing humoral immune responses, which elicited the higher titer of IgG antibodies than the other recombinant protein vaccines[21,32]. On the one hand, TT-P2 epitope as an endogenous adjuvant could be recognized by antigen-presenting cells (APCs) and presented to effector T lymphocytes, which further stimulated the proliferation and differentiation of B cells secreting antibodies against the subunit antigen[33]. On the other hand, CpG 1826 plus alum dual adjuvant could synergistically enhance the humoral immune responses induced by subunit vaccines, which was in accordance with the previous studies[12,16,34]. CpG 1826 as a pathogen-associated molecular pattern (PAMP) recognized by Toll-like receptor 9 (TLR9) could stimulate the activation and proliferation of B cells by upregulating the expression of co-stimulatory molecules such as CD40, CD80 and CD86[34,35]. Especially after the boost immunization, the body’s accelerated immune responses to the proteins led to the significantly increasing level of serum IgG antibodies (Figure 6).
The NAbs effectively neutralized the virus were considered as key indicator to assess the protective efficacy of vaccines. Our experimental evidence showed that Sot/CA subunit vaccine could induce higher titer of NAbs, which strongly correlated with the high level of IgG antibodies (Figure 8). Comparatively, the level of NAbs induced by So protein without TT-P2 epitope was slightly weaker, suggesting that TT-P2 epitope as an endogenous adjuvant seldom suppressed the recognition of RBD neutralizing epitopes and effectively increased the level of NAbs by strengthening the interactions of T-B cells[21,36]. Moreover, CpG 1826 plus alum dual adjuvant synergistically enhanced the GMT of NAbs induced by vaccines, which was consistent with previous studies[15,16,37]. In addition, cross-neutralizing activity against XBB.1.5 variant in mice sera induced by subunit vaccines was also observed, which was a recombinant variant of two descendents from the BA.2 lineage[38]. More importantly, XBB.1.5 and BA.4/5 variants shared many neutralizing epitopes, we speculated that mice serum immunized with subunit vaccines also exhibited cross-neutralizing activity against BA.4/5, BF.4.6 and BQ.1 variants[37,39]. However, the level of NAbs induced by Sot/CA subunit vaccine were not as good as mRNA vaccine. This might be explained by the mRNA constantly expressed SARS-CoV-2 protein antigens in the host cells, which maintaining for a long time and promoting a robust germinal center (GC) responses[40]. In summary, Sot protein formulated with CpG 1826 plus alum dual adjuvant showed great humoral immunogenicity, which induced the high level of IgG antibodies and cross NAbs against BA.2 and XBB.1.5 variants.
In addition to the antibody responses, cellular immune responses also played a vital role in defensing against SARS-CoV-2 infection. Previous studies reported that cellular immunity against SARS-CoV-2 maintained for 6 months after the vaccination, indicating the cellular immune responses might have a long-term effect on fighting against COVID-19[41-43]. ELISpot results showed that Sot/CA subunit vaccine significantly activated effector T lymphocytes secreting IFN-γ and IL-4, which were significantly higher than those by the other vaccines[21] (Figure 9A and 9B). For one thing, TT-P2 epitope as an endogenous adjuvant recognized by the most HLA-DR molecules could effectively activate T lymphocytes, which remarkably promoted the secretion of IFN-γ and IL-4[10,11]. For another, CpG 1826 plus alum dual adjuvant simultaneously promoted Th1 and Th2-type immune responses that secretion of IFN-γ and IL-4, which was consistent with the other studies[12,15]. TLR9 activated by CpG 1826 could trigger the signal transduction dependent on myeloid differentiation primary responses protein 88 (MyD88) and subsequently enhance the expression of downstream transcription factor such as nuclear factor-kappa B (NF-κB), which further promoted the secretion of IFN-γ and IL-4[44,45]. The secretion of IFN-γ was closely related with Th1-type immune response, while IL-4 was the major cytokine associated with Th2-type immune response[46,47]. Combined the decreased IFN-γ/IL-4 SFCs ratio (Figure 9C) with the increased IgG1/IgG2a ratio (Figure 7C), our results illustrated that alum adjuvant preferentially induced Th2-type immune responses[21,48]. Conversely, the increased IFN-γ/IL-4 SFCs ratio (Figure 9C) and the decreased IgG1/IgG2a ratio (Figure 7C) suggested that CpG 1826 and alum dual adjuvant induced predominantly Th1-type immune response, which was crucial to successfully control of SARS-CoV-2 replication[47,49]. These results might be explained by the excellent combination of CpG 1826 with alum adjuvant improved the stability of CpG 1826, which further exhibited the immune activation of CpG 1826 adjuvant. In summary, Sot protein formulated with CpG 1826 and alum dual adjuvant was of great cellular immunogenicity, which induced the high level of effector T lymphocytes secreting the IFN-γ and IL-4.
There were also some limitations about this study. Firstly, Sot subunit antigen was monomeric RBD, with small molecular weight and weaker immunogenicity. The GMT of NAbs induced in mice was not as excellent as the polymeric protein vaccines[50,51]. Secondly, there were many challenges about the clinical application of CpG 1826 adjuvant, and more studies would be performed to verify the safety profile and molecular mechanisms of the CpG 1826 plus alum dual adjuvant system. Finally, the titer of NAbs induced by the subunit vaccines was just evaluated by the cellular level. In the future study, we would further investigate the protective efficacy of the Sot subunit vaccine formulated with CpG 1826 plus alum dual adjuvant through the virus-challenging experiments in the suitable animal models, such as hACE2-transgenic mice and so on.
In conclusion, Sot recombinant protein, containing TT-P2 epitope, formulated with CpG 1826 plus alum dual adjuvant induced the robust cellular and humoral immune responses and showed great cross-neutralizing activity against BA.2 and XBB.1.5 variants, which should be considered as vaccine candidate and further investigated in the subsequent pre-clinical studies. Given the current epidemiology with the predominance of BA.2-derived subvariants, our study provided a scientific basis for the development of BA.2 variant vaccines and references for the adjuvant application of subunit vaccines.
doi: 10.3967/bes2024.129
Immunogenicity Evaluation of a SARS-CoV-2 BA.2 Subunit Vaccine Formulated with CpG 1826 plus alum Dual Adjuvant
-
Abstract:
Objective The present study aimed to evaluate the immunogenicity of BA.2 variant receptor binding domain (RBD) recombinant protein formulated with CpG 1826 plus alum dual adjuvant. Methods The BA.2 variant RBD (residues 308-548) fusing TT-P2 epitope was obtained from prokaryotic expression system, purification technology and dialysis renaturation, which was designated as Sot protein. The soluble Sot protein formulated with CpG 1826 plus alum dual adjuvant was designated as Sot/CA subunit vaccine and then the BALB/c mice were intramuscularly administrated with two doses of the Sot/CA subunit vaccine at 14-day interval (day 0 and 14). On day 28, the number of effector T lymphocytes secreting IFN-γ and IL-4 in mice spleen were determined by enzyme-linked immunospot (ELISpot) assay. The serum IgG, IgG1 and IgG2a antibodies were examined by enzyme-linked immunosorbent assay (ELISA). In addition, the level of neutralizing antibodies (NAbs) induced by Sot/CA subunit vaccine was also evaluated by the microneutralization assay. Results The high-purity soluble Sot protein with antigenicity was successfully obtained by the prokaryotic expression, protein purification and dialysis renaturation. The Sot/CA subunit vaccine induced a high level of IgG antibodies and NAbs, which were of cross-neutralizing activity against SARS-CoV-2 BA.2 and XBB.1.5 variants. Meanwhile, Sot/CA subunit vaccine also induced a high level of effector T lymphocytes secreting IFN-γ (635.00 ± 17.62) and IL-4 (279.20 ± 13.10), respectively. Combined with a decreased IgG1/IgG2a ratio in the serum, which indicating Sot/CA subunit vaccine induced a Th1-type predominant immune response. Conclusion The Sot protein formulated with CpG 1826 plus alum dual adjuvant showed that the excellent cellular and humoral immunogenicity, which provided a scientific basis for the development of BA.2 variant subunit vaccines and references for the adjuvant application of subunit vaccines. -
Key words:
- SARS-CoV-2 /
- RBD /
- Subunit vaccine /
- Adjuvant /
- Immunogenicity
注释:1) CONFLICT OF INTEREST: -
Figure 3. Expression and purification of recombinant proteins. (A) SDS-PAGE analysis of the recombinant So protein: Lane M, protein marker; lane 1, non-induced bacteria; lane 2, induced bacteria; lane 3, soluble fraction; lane 4, IBs; lane 5, denatured So protein; lane 6, So protein purified by IEC; lane 7, So protein purified by Ni-NTA. (B) SDS-PAGE analysis of the recombinant Sot protein: Lane M, protein marker; lane 1, non-induced bacteria; lane 2, induced bacteria; lane 3, soluble fraction; lane 4, IBs; lane 5, denatured Sot protein; lane 6, Sot protein purified by IEC; lane 7, Sot protein purified by Ni-NTA. SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; IBs, inclusion bodies; IEC, ion-exchange chromatography.
Figure 4. Antigenicity analysis of the recombinant proteins: (A) WB assay of the proteins: Lane M, protein marker; lane 1, So protein; lane 2, Sot protein. (B) Absorbance at 450 nm of ELISA. WB, Western blotting; ELISA, enzyme-linked immunosorbent assay; HD, healthy donors; COVID-19, coronavirus disease 2019.
Figure 6. The titer of specific IgG antibodies in immunized mice. (A) The titer of IgG antibodies on day 14. (B) The titer of IgG antibodies on day 28. dpi, day post immunization; GMT, geometric mean titer. The data were analyzed to evaluate significant differences by Two-way ANOVA. Asterisk (*) represents the difference between the experimental groups. ns (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 7. Analysis of IgG antibody isotypes in immunized mice. (A) The titer of IgG1 antibodies. (B) The titer of IgG2a antibodies. (C) The IgG1/IgG2a ratios in mice serum. GMT, geometric mean titer. The data were analyzed to evaluate significant differences by Two-way ANOVA. Asterisk (*) represents the difference between the experimental groups. ns, P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001.
Figure 8. The titer of NAbs in mice serum. (A) The titer of NAbs against BA.2 variant. (B) The titer of NAbs against XBB.1.5 variant. NAbs, neutralizing antibodies; GMT, geometric mean titer. The data were analyzed by Two-way ANOVA to evaluate significant differences. Asterisk (*) represents the difference between the experimental groups. ns, no significance; P > 0.05; *P < 0.05.
Figure 9. The SFCs secreting IFN-γ and IL-4 in mice spleen. (A) The SFCs of IFN-γ. (B) The SFCs of IL-4. (C) The ratios of IFN-γ/IL-4 SFCs. SFCs, spot-forming cells. The data were analyzed by Two-way ANOVA to evaluate significant differences and expressed as mean ± SD. Asterisk (*) represents the difference between the experimental groups. ns, no significance; P > 0.05, *P < 0.05, ****P < 0.0001.
Table 1. Animal grouping and vaccination regimen of mice
Group Adjuvant Route Dose Interval N# Adjuvant CpG 1826+alum i.m. 100 μL/50 μg 0, 14 5 Adjuvant CpG 1826 i.m. 100 μL/50 μg 0, 14 5 Adjuvant alum i.m. 100 μL/50 μg 0, 14 5 So/alum alum i.m. 40 μg/50 μg 0, 14 5 Sot/alum alum i.m. 40 μg/50 μg 0, 14 5 So/CpG 1826 CpG 1826 i.m. 40 μg/50 μg 0, 14 5 Sot/CpG 1826 CpG 1826 i.m. 40 μg/50 μg 0, 14 5 So/CA CpG 1826+alum i.m. 40 μg/50 μg 0, 14 5 Sot/CA CpG 1826+alum i.m. 40 μg/50 μg 0, 14 5 Note. i.m.: intramuscular injection; N#, number of mice. -
[1] World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. 2024. https://dashboards-dev.sprinklr.com/. [2] Uraki R, Kiso M, Iida S, et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA. 2. Nature, 2022; 607, 119-27 . DOI: 10.1038/s41586-022-04856-1. [3] Shrestha LB, Foster C, Rawlinson W, et al. Evolution of the SARS-CoV-2 omicron variants BA. 1 to BA. 5: implications for immune escape and transmission. Rev Med Virol, 2022; 32, e2381. doi: 10.1002/rmv.2381 [4] Li P, Faraone JN, Hsu CC, et al. Characteristics of JN. 1-derived SARS-CoV-2 subvariants SLip, FLiRT, and KP. 2 in neutralization escape, infectivity and membrane fusion. https://doi.org/10.1101/2024.05.20.595020. [2024-05-21]. [5] Tamura T, Ito J, Uriu K, et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun, 2023; 14, 2800. doi: 10.1038/s41467-023-38435-3 [6] Muik A, Lui BG, Bacher M, et al. Omicron BA. 2 breakthrough infection enhances cross-neutralization of BA. 2.12. 1 and BA. 4/BA. 5. Sci Immunol, 2022; 7, eade2283. doi: 10.1126/sciimmunol.ade2283 [7] World Health Organization. Draft landscape and tracker of COVID-19 candidate vaccines. 2024. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [8] Pollet J, Chen WH, Strych U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv Drug Delivery Rev, 2021; 170, 71−82. doi: 10.1016/j.addr.2021.01.001 [9] Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 2020; 181, 281-92. e6. [10] Panina-Bordignon P, Tan A, Termijtelen A, et al. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol, 1989; 19, 2237−42. doi: 10.1002/eji.1830191209 [11] Cai H, Chen MS, Sun ZY, et al. Self-adjuvanting synthetic antitumor vaccines from MUC1 glycopeptides conjugated to T-cell epitopes from tetanus toxoid. Angew Chem Int Ed, 2013; 52, 6106−10. doi: 10.1002/anie.201300390 [12] Wu F, Yuan XY, Li J, et al. The co-administration of CpG-ODN influenced protective activity of influenza M2e vaccine. Vaccine, 2009; 27, 4320−4. doi: 10.1016/j.vaccine.2009.04.075 [13] Pun PB, Bhat AA, Mohan T, et al. Intranasal administration of peptide antigens of HIV with mucosal adjuvant CpG ODN coentrapped in microparticles enhances the mucosal and systemic immune responses. Int Immunopharmacol, 2009; 9, 468−77. doi: 10.1016/j.intimp.2009.01.012 [14] Kumar S, Jones TR, Oakley MS, et al. CpG oligodeoxynucleotide and Montanide ISA 51 adjuvant combination enhanced the protective efficacy of a subunit malaria vaccine. Infect Immun, 2004; 72, 949−57. doi: 10.1128/IAI.72.2.949-957.2004 [15] Zhang YT, Zheng XT, Sheng W, et al. Alum/CpG adjuvanted inactivated COVID-19 vaccine with protective efficacy against SARS-CoV-2 and variants. Vaccines (Basel), 2022; 10, 1208. doi: 10.3390/vaccines10081208 [16] Nanishi E, Borriello F, O’Meara TR, et al. An aluminum hydroxide: CpG adjuvant enhances protection elicited by a SARS-CoV-2 receptor binding domain vaccine in aged mice. Sci Transl Med, 2022; 14, eabj5305. doi: 10.1126/scitranslmed.abj5305 [17] Deng Y, Lan JM, Bao LL, et al. Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus. Emerg Microbes Infect, 2018; 7, 60. [18] Gong MQ, Zhou J, Yang CT, et al. Insect cell-expressed hemagglutinin with CpG oligodeoxynucleotides plus alum as an adjuvant is a potential pandemic influenza vaccine candidate. Vaccine, 2012; 30, 7498−505. doi: 10.1016/j.vaccine.2012.10.054 [19] Xiao TY, Liu HC, Li XQ, et al. Immunological evaluation of a novel mycobacterium tuberculosis antigen Rv0674. Biomed Environ Sci, 2019; 32, 427−37. [20] Chen WH, Pollet J, Strych U, et al. Yeast-expressed recombinant SARS-CoV-2 receptor binding domain RBD203-N1 as a COVID-19 protein vaccine candidate. Protein Expression Purif, 2022; 190, 106003. doi: 10.1016/j.pep.2021.106003 [21] Su QD, Zou YN, Yi Y, et al. Recombinant SARS-CoV-2 RBD with a built in T helper epitope induces strong neutralization antibody response. Vaccine, 2021; 39, 1241−7. doi: 10.1016/j.vaccine.2021.01.044 [22] He YX, Zhou YS, Liu SW, et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun, 2004; 324, 773−81. doi: 10.1016/j.bbrc.2004.09.106 [23] Ai JW, Wang X, He XY, et al. Antibody evasion of SARS-CoV-2 Omicron BA. 1, BA. 1.1, BA. 2, and BA. 3 sub-lineages. Cell Host Microbe, 2022; 30, 1077−83. doi: 10.1016/j.chom.2022.05.001 [24] Hu YM, Huang SJ, Chu K, et al. Safety of an Escherichia coli-expressed bivalent human papillomavirus (types 16 and 18) L1 virus-like particle vaccine: an open-label phase I clinical trial. Hum Vaccines Immunother, 2014; 10, 469−75. doi: 10.4161/hv.26846 [25] Zhu FC, Zhang J, Zhang XF, et al. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet, 2010; 376, 895−902. doi: 10.1016/S0140-6736(10)61030-6 [26] Lee SH, Carpenter JF, Chang BS, et al. Effects of solutes on solubilization and refolding of proteins from inclusion bodies with high hydrostatic pressure. Protein Sci, 2006; 15, 304−13. doi: 10.1110/ps.051813506 [27] Fraga TR, Chura-Chambi RM, Gonçales AP, et al. Refolding of the recombinant protein OmpA70 from Leptospira interrogans from inclusion bodies using high hydrostatic pressure and partial characterization of its immunological properties. J Biotechnol, 2010; 148, 156−62. doi: 10.1016/j.jbiotec.2010.04.007 [28] Deng TT, Li TT, Chen GG, et al. Characterization and immunogenicity of SARS-CoV-2 spike proteins with varied glycosylation. Vaccine, 2022; 40, 6839−48. doi: 10.1016/j.vaccine.2022.09.057 [29] Shajahan A, Supekar NT, Gleinich AS, et al. Deducing the N-and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology, 2020; 30, 981−8. doi: 10.1093/glycob/cwaa042 [30] Huang HY, Liao HY, Chen XR, et al. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci Transl Med, 2022; 14, eabm0899. doi: 10.1126/scitranslmed.abm0899 [31] De Marco Verissimo C, Corrales JL, Dorey AL, et al. Production of a functionally active recombinant SARS-CoV-2 (COVID-19) 3C-like protease and a soluble inactive 3C-like protease-RBD chimeric in a prokaryotic expression system. Epidemiol Infect, 2022; 150, e128. doi: 10.1017/S0950268822001078 [32] Merkuleva IA, Shcherbakov DN, Borgoyakova MB, et al. Comparative immunogenicity of the recombinant receptor-binding domain of protein S SARS-CoV-2 obtained in prokaryotic and mammalian expression systems. Vaccines (Basel), 2022; 10, 96. doi: 10.3390/vaccines10010096 [33] Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol, 1999; 17, 593−623. doi: 10.1146/annurev.immunol.17.1.593 [34] Davis H L, Weeranta R, Waldschmidt T J, et al. CpG DNA is a potent enhancer of specific immunity in mice immunized with recombinant hepatitis B surface antigen. J Immunol, 1998; 160, 870−6. doi: 10.4049/jimmunol.160.2.870 [35] Krieg AM, Yi AK, Matson S, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature, 1995; 374, 546−9. doi: 10.1038/374546a0 [36] Kumar A, Arora R, Kaur P, et al. "Universal" T helper cell determinants enhance immunogenicity of a Plasmodium falciparum merozoite surface antigen peptide. J Immunol, 1992; 148, 1499−505. doi: 10.4049/jimmunol.148.5.1499 [37] Thimmiraju SR, Adhikari R, Villar MJ, et al. A recombinant protein XBB. 1.5 RBD/Alum/CpG vaccine elicits high neutralizing antibody titers against omicron subvariants of SARS-CoV-2. Vaccines (Basel), 2023; 11, 1557. doi: 10.3390/vaccines11101557 [38] Channabasappa NK, Niranjan AK, Emran TB. SARS-CoV-2 variant omicron XBB. 1.5: challenges and prospects-correspondence. Int J Surg, 2023; 109, 1054−5. doi: 10.1097/JS9.0000000000000276 [39] Muik A, Lui B G, Bacher M, et al. Exposure to BA. 4/5 S protein drives neutralization of Omicron BA. 1, BA. 2, BA. 2.12. 1, and BA. 4/5 in vaccine-experienced humans and mice. Sci Immunol, 2022; 7, eade9888. doi: 10.1126/sciimmunol.ade9888 [40] Lederer K, Castaño D, Atria DG, et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity, 2020; 53, 1281-95. e5. [41] Kato H, Miyakawa K, Ohtake N, et al. Vaccine-induced humoral response against SARS-CoV-2 dramatically declined but cellular immunity possibly remained at 6 months post BNT162b2 vaccination. Vaccine, 2022; 40, 2652−5. doi: 10.1016/j.vaccine.2022.03.057 [42] Steiner S, Schwarz T, Corman VM, et al. Reactive T cells in convalescent COVID-19 patients with negative SARS-CoV-2 antibody serology. Front Immunol, 2021; 12, 687449. doi: 10.3389/fimmu.2021.687449 [43] Feng CQ, Shi JR, Fan QH, et al. Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat Commun, 2021; 12, 4984. doi: 10.1038/s41467-021-25312-0 [44] Kumagai Y, Takeuchi O, Akira S. TLR9 as a key receptor for the recognition of DNA. Adv Drug Delivery Rev, 2008; 60, 795−804. doi: 10.1016/j.addr.2007.12.004 [45] He WX, Zhang YQ, Zhang J, et al. Cytidine-phosphate-guanosine oligonucleotides induce interleukin-8 production through activation of TLR9, MyD88, NF-κB, and ERK pathways in odontoblast cells. J Endod, 2012; 38, 780−5. doi: 10.1016/j.joen.2012.02.026 [46] Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol, 1989; 7, 145−73. doi: 10.1146/annurev.iy.07.040189.001045 [47] Qi M, Zhang XE, Sun XX, et al. Intranasal nanovaccine confers homo- and hetero-subtypic influenza protection. Small, 2018; 14, e1703207. doi: 10.1002/smll.201703207 [48] He P, Zou YN, Hu ZY. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum Vaccines Immunother, 2015; 11, 477−88. doi: 10.1080/21645515.2014.1004026 [49] Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol, 2020; 38, 1−9. [50] Liang JG, Su DM, Song TZ, et al. S-trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat Commun, 2021; 12, 1346. doi: 10.1038/s41467-021-21634-1 [51] Leal L, Pich J, Ferrer L, et al. Safety and immunogenicity of a recombinant protein RBD fusion heterodimer vaccine against SARS-CoV-2. NPJ Vaccines, 2023; 8, 147. doi: 10.1038/s41541-023-00736-5 -




 下载:
下载:



 Quick Links
Quick Links