-
Nasopharyngeal carcinoma (NPC) is a common malignant tumor of the head and neck[1]. Although significant results have been achieved in the comprehensive treatment of NPC[2,3], the pathogenesis and progression of NPC remain unclear. Thus, exploring NPC pathogenesis to identify novel molecular targets will help identify new treatment strategies.
The tumor microenvironment plays an important role in malignant tumor progression. Tumor-associated macrophages (TAMs) are infiltrating macrophages in tumor tissues that participate in the regulation of the growth, invasion, and metastasis of various tumors by secreting a variety of cytokines. TAMs are heterogeneous and exhibit two differentiation types (M1 and M2)[4,5]. M2 macrophages play an important role in promoting tumor growth and immunosuppression[6]. M2 macrophages are associated with the progression and prognosis of NPC[7,8].
An increasing number of studies have shown that microRNAs (miRNAs) play important roles in macrophage differentiation and activity maintenance[9,10]. Moreover, the miR-34 family has been studied in various tumors, including miR-34a, miR-34b, and miR-34c[11]. miR-34a deletion affects M2 macrophages and further affects doxorubicin resistance in uterine leiomyoma[12]. Adipocyte-secreted exosomal miR-34a prevents M2 macrophages from promoting obesity-induced adipose inflammation[13]. SRT1720 suppresses the M1 polarization of macrophages by targeting miR-34b-5p to reduce mitochondrial damage in tubular epithelial cells[14]. However, few studies have investigated the correlation between miR-34c and macrophages in tumors. Thus, this study aimed to explore whether miR-34c regulates M2 macrophages to affect NPC progression and elucidate the underlying mechanisms.
-
THP-1 cells (YaJi Biotech, Shanghai, China) in the logarithmic growth phase were treated with phorbol 12-myristate 13-acetate (PMA, 100 ng/mL, Beyotime, Shanghai, China) for 24 h to induce M0 macrophages. Then, M0 cells were treated with recombinant lipopolysaccharide (LPS,5 μL, 1:1,000, Beyotime, Shanghai, China) and human interferon-γ (IFN-γ, 5 μL, 1:1,000, Beyotime, Shanghai, China) to induce the M1 phenotype, and recombinant human interleukin-4 (IL-4, 5 μL, 1:500, Beyotime, Shanghai, China) and human IL-13 (5 μL, 1:500, Beyotime, Shanghai, China) were employed to induce the M2 phenotype. An optical microscope (Leica Microsystems, Wetzlar, Germany) was used to observe M1 and M2 macrophages.
-
The induced M2 macrophages were transfected with the normal control (NC) mimic or miR-34c-3p mimic using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s protocol. The transfected cells were harvested after 48 h to 72 h.
-
The proportions of M1 and M2 macrophages were determined using flow cytometry (BD Biosciences, San Jose, CA, USA). Briefly, forward scatter (FSC) and side scatter (SSC) gates were used to remove the cell debris and clumped cells to circle the target cell population. Then, the FSC-area (FSC-A) and FSC-height (FSC-H) gates were used to circle the single-cell population, and F4/80 was used to circle the macrophages. CD86 was used to circle M1 macrophages, and CD206 was used to circle M2 macrophages.
-
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into complementary DNA using a reverse transcription kit (Aidlab Biotech, Beijing, China). The qRT-PCR was performed using an ABI-7500 Real-Time PCR System (Applied Biosystems, Warrington, UK). The data were analyzed using the 2-∆∆Ct method, and β-actin was used for normalization. The primers used were listed in Table 1.
Table 1. The primers of genes
Gene Sequences (5′–3′) iNOS (forward) TTCAGTATCACAACCTCAGCAAG iNOS (reverse) TGGACCTGCAAGTTAAAATCCC Arg-1 (forward) GTGGAAACTTGCATGGACAAC Arg-1 (reverse) AATCCTGGCACATCGGGAATC IL-10 (forward) GACTTTAAGGGTTACCTGGGTTG IL-10 (reverse) TCACATGCGCCTTGATGTCTG IL-6 (forward) ACTCACCTCTTCAGAACGAATTG IL-6 (reverse) CCATCTTTGGAAGGTTCAGGTTG TNF-α (forward) CCTCTCTCTAATCAGCCCTCTG TNF-α (reverse) GAGGACCTGGGAGTAGATGAG SLC7A11 (forward) TCTCCAAAGGAGGTTACCTGC SLC7A11 (reverse) AGACTCCCCTCAGTAAAGTGAC β-actin (forward) AGCGAGCATCCCCCAAAGTT β-actin (reverse) GGGCACGAAGGCTCATCATT -
The protein content was determined using a bicinchoninic acid (BCA) protein assay kit (Vazyme, Nanjing, China). The proteins were then separated, transferred to polyvinylidene fluoride (PVDF) membranes, and maintained with 5% non-fat milk for 1 h at room temperature ranged from 20 to 25. The membranes were then incubated with primary antibodies specific for S°CLC7A11 (26864-1-AP, 1:1,000; Proteintech, Chicago, IL, USA), β-actin (GB11001, 1:2,500; Servicebio, Wuhan, China), and secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (ab6721, 1:2,000; Abcam, Cambridge, MA, USA). Finally, an enhanced chemiluminescence kit (Biosharp, Beijing, China) was used, and optical density was analyzed using Image-Pro Plus 6.0 software (MediaCybernetics, Bethesda, MD, USA). β-actin acted as the internal control.
-
The Cell Counting Kit-8 (CCK8) assay was performed to examine the viability of NPC cells as described previously[15], with minor modifications. Briefly, cells were seeded into a 96-well plate at a density of 5,000 cells/well and incubated at 37 °C and 5% CO2 for 24 h. Then, the CCK8 reagent (Sigma, Saint Louis, MO, USA) was added to each well and incubated with cells for 4 h. The optical density of each well was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
-
Cell migration and invasion were assessed as previously described[16]. NPC cells were resuspended in serum-free dulbecco’s modified eagle medium (DMEM) and inoculated into the upper layer of the Transwell plate. In the migration experiments, the upper layer of the transwell chamber was not pretreated, and the serum-free supernatant of NPC cells was added to the lower layer. In the invasion experiments, the upper layer of the Transwell chamber was pre-coated with matrix glue, and DMEM containing 15% fetal bovine serum (FBS) was added to the lower layer. The experiments were terminated after a certain period of culture (16 h for migration and 24 h for invasion). The cells in the upper layer of the transwell chamber were wiped with cotton swabs, whereas the cells in the lower layer were not removed. The cells were stained with 1% crystal purple and observed under a microscope.
-
The induced M2 macrophages were co-cultured with NPC cells in the logarithmic growth phase at a ratio of 1:1 and cultured for 24–48 h for the following experiments.
-
The SLC7A11-3′UTR containing wild or mutant binding sites was cloned into the psiCHECKTM-2 vector (Genepharma, Shanghai, China), respectively. Then HEK-293 T cells were co-transfected with miR-34c-3p mimic and SLC7A11-3′UTR psiCHECKTM-2 plasmid; mimic NC was the control. The cells were harvested and washed two times with phosphate buffer saline (PBS). Luciferase activity was measured using a dual-luciferase reporter assay system (E1910, Promega, Beijing, China).
-
NPC was induced by the right underarm injection of 0.2 mL 5-8F cell suspension (5 × 1010/L) into male BALB/c mice (3–5 weeks old), and mice injected with normal saline were used as the control group. The model mice were then injected with miR-34c-3p mimic or mimic NC via the tail vein. The tumor growth curves of the mice were assessed every 3 d, and the tumor weight was examined after 15 d. All animal experiments were approved by the Ethics Committee of Henan Provincial People’s Hospital.
-
After fixation, embedding, deparaffinization, and rehydration, the tumor tissues were cut into 4-μm sections and heated for antigen retrieval. Sections were incubated with 3% hydrogen peroxidase in methanol for 5 min and then incubated with primary polyclonal antibodies against CD86 (1:100, Cell Signaling Technology, Beverly, MA, USA) and CD206 (1:200, Cell Signaling Technology, Beverly, MA, USA) for 15 min at room temperature. The sections were then incubated with a horseradish peroxidase-conjugated secondary antibody for 30 min. Finally, the sections were stained with hematoxylin and observed under an optical microscope.
-
After fixation with 4% formaldehyde at 20 °C for 30 min, the tumor tissues were embedded in paraffin and cut into 5-μm sections. The sections were stained with hematoxylin and eosin (Solarbio, Wuhan, China) for 10 min. Images were captured under a light microscope (×200, OLYMPUS DP70, Tokyo, Japan), and pathological changes in the tissues and infiltration of inflammatory cells were observed.
-
Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Inc., San Diego, CA, USA). Differences between groups were analyzed using the unpaired Student’s t-test or one-way analysis of variance (ANOVA), followed by Tukey’s post-hoc tests. P values less than 0.05 were considered significant.
-
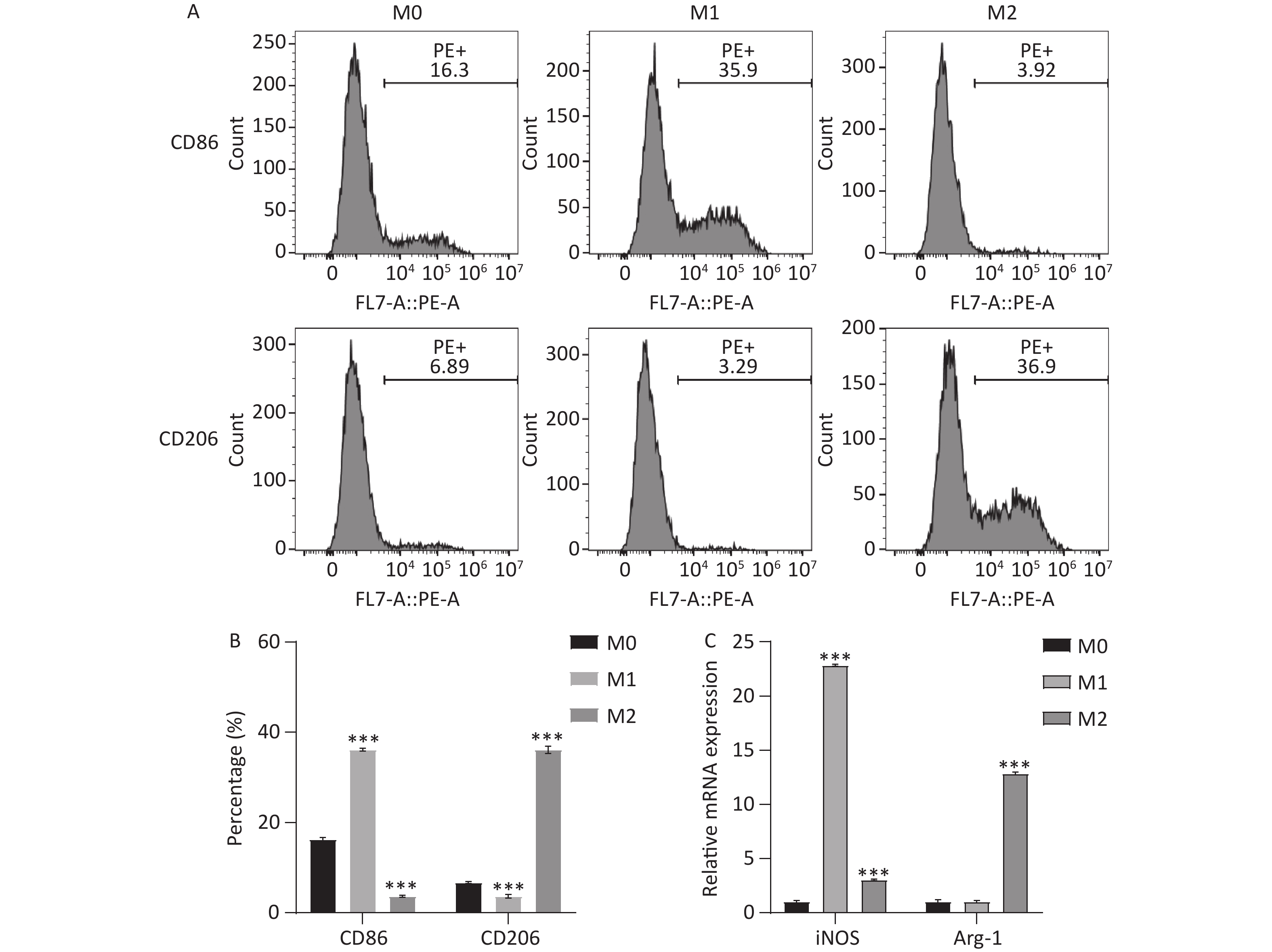
First, THP-1 cells were treated with PMA to induce M0 cells, and M0 cells were further treated with different cytokines to induce M1 and M2 phenotype, respectively. Flow cytometry and qRT-PCR analysis revealed that, compared with M0 cells, cells with the M1 phenotype exhibited higher expression of CD86 and iNOS, whereas cells with the M2 phenotype exhibited higher expression of CD206 and Arg-1 (Figure 1A–C). These results suggested the successful establishment of M1 and M2 macrophages.

Figure 1. Induction of M1 and M2 macrophages. THP-1 cells were treated with PMA to induce M0 cells, and M0 cells were further treated with different cytokines to induce M1 and M2 phenotype, respectively. (A, B) Flow cytometry was performed to detect the expression of CD86 and CD206. (C) qRT-PCR was performed to examine iNOS and Arg-1 expression. ***P < 0.001.
-
Compared with M0 cells, M1 macrophages exhibited no obvious alteration in miR-34c-3p expression, whereas M2 macrophages exhibited a significant decrease in miR-34c-3p expression (Figure 2A). Therefore, M2 macrophages were used in subsequent experiments. miR-34c-3p mimic was used to overexpress miR-34c-3p in M2 macrophages (Figure 2B). Flow cytometry and qRT-PCR analyses revealed that, compared with the mimic NC, the miR-34c-3p mimic significantly upregulated CD86 and iNOS expression while downregulating CD206 and Arg-1 expression (Figure 2C–E). Moreover, increased tumor necrosis factor-α (TNF-α), IL-6, and decreased IL-10 were observed in M2 macrophages transfected with miR-34c-3p mimic (Figure 2F). These results suggested that overexpression of miR-34c-3p markedly suppressed M2 polarization in macrophages.

Figure 2. miR-34c-3p inhibits M2 macrophage polarization. (A) qRT-PCR was performed to examine the expression of miR-34c-3p in different cells. (B) Induced M2 macrophages were transfected with miR-34c-3p mimic or mimic NC, and qRT-PCR was performed to examine the expression of miR-34c-3p. (C, D) Flow cytometry was performed to detect the expression of CD86 and CD206. qRT-PCR was employed to examine the expression of iNOS, Arg-1 (E), and TNF-α, IL-6, and IL-10 (F). ** P < 0.01, *** P < 0.001.
-
Next, M2 macrophages transfected with miR-34c-3p mimic were co-cultured with NPC cells (5-8F and C666-1 cells). CCK8 and transwell assays revealed that M2 macrophages significantly promoted the proliferation, migration, and invasion of 5-8F cells compared to M0 cells, whereas M2 macrophages transfected with the miR-34c-3p mimic markedly downregulated the increased proliferation, migration, and invasion of 5-8F cells co-cultured with M2 macrophages transfected with the mimic NC (Figure 3). Similar results were observed in C666-1 cells (Figure 4). Based on the results illustrated in Figures 2–4, we concluded that the overexpression of miR-34c-3p inhibited NPC progression by inhibiting the M2 polarization of macrophages in vitro.

Figure 3. miR-34c-3p-mediated M2 macrophages suppresses proliferation, migration, and invasion in 5-8F cells. M2 macrophages transfected with the miR-34c-3p mimic or NC mimic were co-cultured with 5-8F cells. (A) The CCK8 assay was used to assess cell proliferation. Transwell assays were performed to assess cell migration (B, C) and invasion (D, E). **P < 0.01, ***P < 0.001.
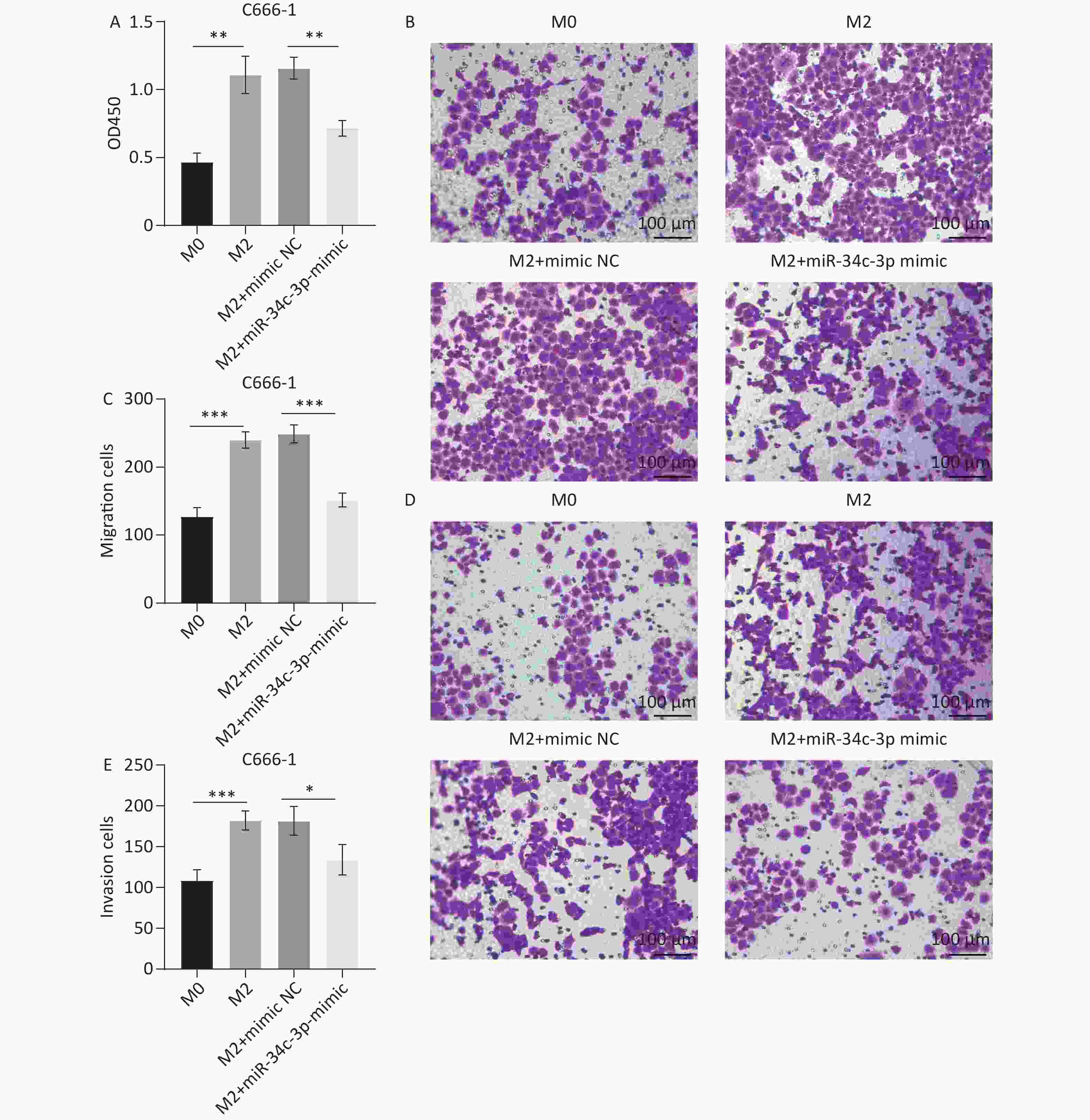
Figure 4. miR-34c-3p-mediated M2 macrophages suppresses proliferation, migration, and invasion in C666-1 cells. M2 macrophages transfected with the miR-34c-3p mimic or NC mimic were co-cultured with C666-1 cells. (A) The CCK8 assay was used to assess cell proliferation. Transwell assays were performed to assess cell migration (B, C) and invasion (D, E). *P < 0.05, **P < 0.01, ***P < 0.001.
-
The predicted specific binding sites between miR-34c-3p and SLC7A11 are shown in Figure 5A, and a dual-luciferase reporter assay confirmed the interaction between miR-34c-3p and SLC7A11 (Figure 5B). The effect of miR-34c-3p on SLC7A11 expression in M2 macrophages was examined using qRT-PCR and western blotting, and the results revealed that the miR-34c-3p mimic significantly reduced SLC7A11 expression compared to the mimic NC (Figure 5C and D). Furthermore, the increase in CD86 expression and the decrease in CD206 expression induced by the miR-34c-3p mimic were reversed by SLC7A11 treatment (Figure 5E and F). These results suggest that overexpression of miR-34c-3p targets SLC7A11 to inhibit M2 polarization in macrophages.
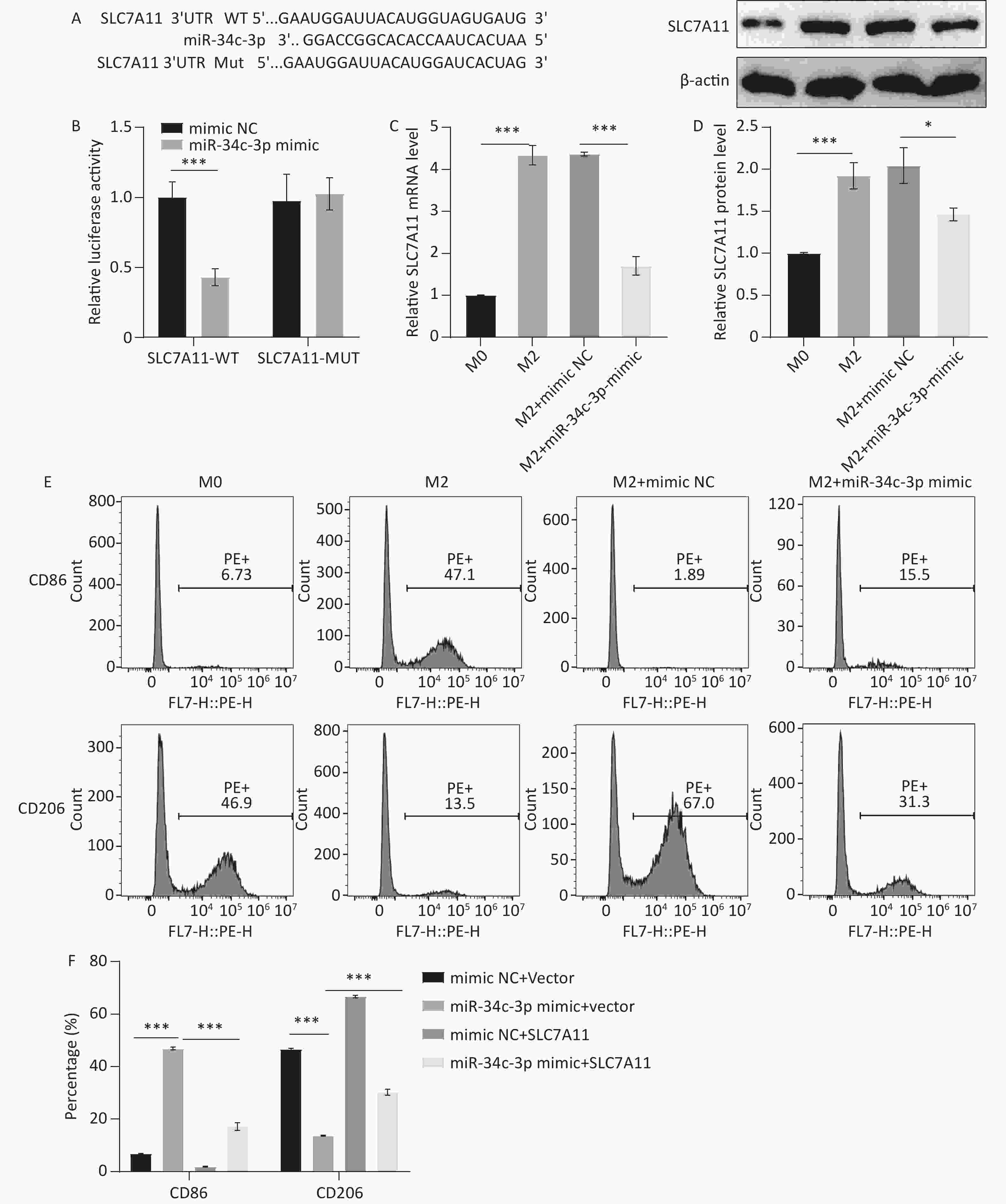
Figure 5. miR-34c-3p targets SLC7A11 to inhibit M2 macrophage polarization. (A) Predicted specific binding sites for miR-34c-3p and SLC7A11. (B) Dual-luciferase reporter assay performed to confirm the interaction between miR-34c-3p and SLC7A11. qRT-PCR (C) and western blotting (D) were performed to examine SLC7A11 mRNA and protein levels. (E, F) Flow cytometry was performed to detect the expression of CD86 and CD206. * P < 0.05, *** P < 0.001.
-
Finally, the miR-34c-3p mimic was injected into the xenograft model for the in vivo experiments. We observed that tumor volume (Figure 6A and B) and weight (Figure 6C) were significantly reduced in the miR-34c-3p mimic group compared to those in the mimic NC group. HE staining revealed that the model group exhibited massive inflammatory infiltration, which was reversed in the miR-34c-3p mimic group (Figure 6D). Furthermore, immunohistochemical staining revealed that the miR-34c-3p mimic elevated CD86 expression and reduced CD206 expression in tumor tissues compared to those in the mimic NC group (Figure 6E and F). These results suggest that the overexpression of miR-34c-3p reduces M2 macrophages and inhibits NPC progression in vivo.
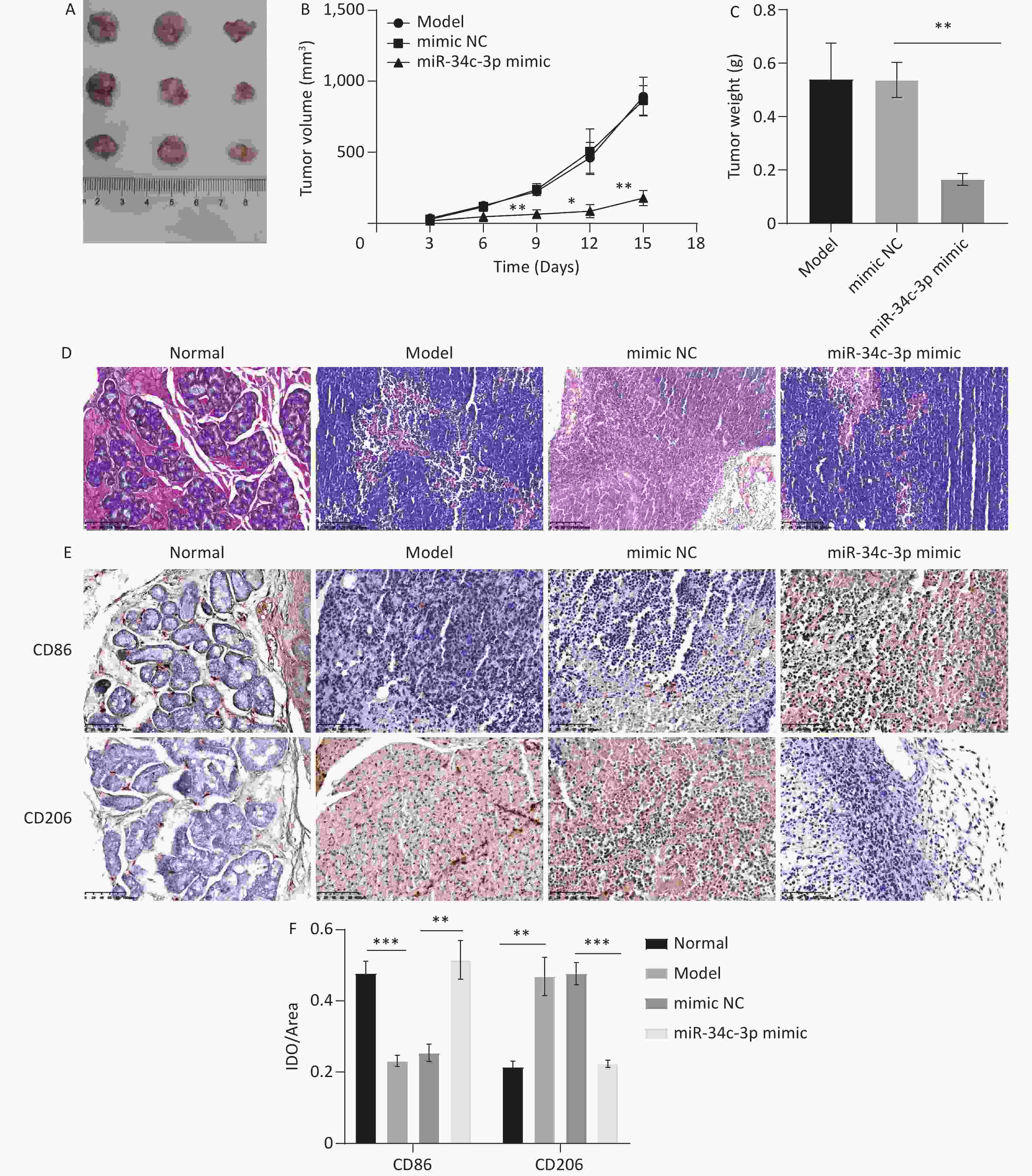
Figure 6. miR-34c-3p blocks NPC progression in vivo. miR-34c-3p mimic was injected into the xenograft model, and the tumor volume (A, B) and weight (C) were measured. (D) HE staining was performed to assess the pathological changes in tumor tissues. (E, F) Immunohistochemical staining was performed to detect CD86 and CD206 expression in tumor tissues. * P < 0.05, ** P < 0.01, *** P < 0.001.
-
miR-34c, including two subtypes, 3p and 5p, plays an important role in tumor development as a tumor suppressor[17,18]. However, most studies of miR-34c in NPC have focused on the subtype 5p. For example, He et al.[19] reported that circCRIM1 promotes NPC progression via the miR-34c-5p/FOSL1 axis, and Huang et al.[20] reported that circ-NOTCH1 promotes aggressive phenotypes of NPC cells by regulating the miR-34c-5p/c-Myc axis. miR-34c-3p is down-regulated in NPC[21], but its role in NPC remains unclear. In this study, we observed that the ectopic expression of miR-34c-3p blocked NPC growth, which corresponded to its role as a tumor suppressor[22,23]. However, the regulatory mechanisms of miR-34c-3p in NPC remain unclear, and further studies are needed.
Macrophages are activated in response to infection or injury to infiltrate inflammatory tissues, recruit lymphocytes, and swallow the invading pathogens and cell debris. Large infiltrated macrophages were observed in the tumor microenvironment, and the tumor-associated macrophages were often divided into two different types: M1 macrophages were activated by LPS, IFN-γ, and TNF-α and secreted inflammatory cytokines such as IL-1β, IL-6, and IL-12, exhibiting an anti-tumor role; M2 macrophages were activated by IL-4 and IL-13 and secreted anti-inflammatory cytokines such as IL-10, CCL18, and CCL22[24]. An increasing number of studies have shown that M2 macrophages are closely involved in the tumor node metastasis classification (TNM) stage, lymph node metastasis, and poor prognosis in patients with NPC[7,25]. M2 macrophages are associated with NPC growth[26]. In this study, we observed that miR-34c-3p expression was significantly decreased in M2 macrophages stimulated with IL-4 and IL-13. miRNAs play a key role in the regulation of macrophage polarization. Downregulation of miR-34a expression affects M2 macrophages and doxorubicin resistance in uterine leiomyomas[12]. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis in colorectal cancer[27]. Gastric cancer-derived exosomal miR-519a-3p promotes liver metastasis by inducing intrahepatic M2 macrophage-mediated angiogenesis[28]. As expected, miR-34c-3p overexpression inhibited M2 macrophage polarization.
To further investigate whether miR-34c-3p affects NPC progression by regulating M2 macrophages, a co-culture system of M2 macrophages and NPC cells was established. We observed that M2 macrophages promoted NPC cell progression, which was reversed by M2 macrophages transfected with the miR-34c-3p mimic, suggesting the involvement of macrophages in the inhibition of NPC progression by miR-34c-3p. In addition to the existence of miRNAs in macrophages, many miRNAs are released into the tumor microenvironment by macrophages through the exosome pathway and absorbed by tumor cells[29,30]. Therefore, in this study, miR-34c-3p may have directly affected NPC cells via exosomes released by macrophages, which requires further investigation.
In addition, we observed that miR-34c-3p targets SLC7A11 to inhibit the M2 polarization of macrophages. SLC7A11 is a ferroptosis-associated gene[31]. Considering the relationship between ferroptosis and macrophage polarization[32], we speculated that miR-34c-3p targeted SLC7A11 to regulate ferroptosis, thus inhibiting M2 polarization of macrophages, which is our next research focus.
In conclusion, this study revealed that miR-34c-3p inhibited NPC progression by inhibiting M2 polarization of macrophages and demonstrated the involvement of ferroptosis in miR-34c-3p-mediated macrophage polarization.
doi: 10.3967/bes2024.136
miR-34c-3p Inhibits Nasopharyngeal Carcinoma Development via Inhibiting M2 Polarization of Macrophages
-
Abstract:
Objective miR-34c-3p is down-regulated in nasopharyngeal carcinoma (NPC). The biological role of miR-34c-3p in NPC and its underlying mechanisms are unknown and were explored in this study. Methods Flow cytometry and immunohistochemical staining were employed to detect cluster of differentiation 86 (CD86) and cluster of differentiation 206 (CD206) expression; quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting were employed to examine mRNA expression and protein levels; cell counting kit-8 (CCK8) and transwell assays were employed to assess cell proliferation, migration, and invasion; and hematoxylin-eosin (HE) staining was employed to assess pathological changes in tumor tissues. Results Our results revealed that the miR-34c-3p mimic markedly inhibited M2 polarization of macrophages by targeting SLC7A11, and M2 macrophages transfected with the miR-34c-3p mimic inhibited the proliferation, migration, and invasion of NPC cells. The in vivo experiments further confirmed that miR-34c-3p mimics blocked tumor growth and reduced inflammatory infiltration in tumor tissues. Conclusion This study provides novel insights into the pathogenesis of NPC and a new treatment strategy. -
Key words:
- miR-34c-3p /
- M2 macrophages /
- Nasopharyngeal carcinoma (NPC) /
- SLC7A11
The authors declare that they have no competing interests in this study.
This study is exempt from ethical review.
&These authors contributed equally to this work.
注释:1) Authors’ Contributions: 2) Competing Interests: 3) Ethics: -
Figure 1. Induction of M1 and M2 macrophages. THP-1 cells were treated with PMA to induce M0 cells, and M0 cells were further treated with different cytokines to induce M1 and M2 phenotype, respectively. (A, B) Flow cytometry was performed to detect the expression of CD86 and CD206. (C) qRT-PCR was performed to examine iNOS and Arg-1 expression. ***P < 0.001.
Figure 2. miR-34c-3p inhibits M2 macrophage polarization. (A) qRT-PCR was performed to examine the expression of miR-34c-3p in different cells. (B) Induced M2 macrophages were transfected with miR-34c-3p mimic or mimic NC, and qRT-PCR was performed to examine the expression of miR-34c-3p. (C, D) Flow cytometry was performed to detect the expression of CD86 and CD206. qRT-PCR was employed to examine the expression of iNOS, Arg-1 (E), and TNF-α, IL-6, and IL-10 (F). ** P < 0.01, *** P < 0.001.
Figure 3. miR-34c-3p-mediated M2 macrophages suppresses proliferation, migration, and invasion in 5-8F cells. M2 macrophages transfected with the miR-34c-3p mimic or NC mimic were co-cultured with 5-8F cells. (A) The CCK8 assay was used to assess cell proliferation. Transwell assays were performed to assess cell migration (B, C) and invasion (D, E). **P < 0.01, ***P < 0.001.
Figure 4. miR-34c-3p-mediated M2 macrophages suppresses proliferation, migration, and invasion in C666-1 cells. M2 macrophages transfected with the miR-34c-3p mimic or NC mimic were co-cultured with C666-1 cells. (A) The CCK8 assay was used to assess cell proliferation. Transwell assays were performed to assess cell migration (B, C) and invasion (D, E). *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 5. miR-34c-3p targets SLC7A11 to inhibit M2 macrophage polarization. (A) Predicted specific binding sites for miR-34c-3p and SLC7A11. (B) Dual-luciferase reporter assay performed to confirm the interaction between miR-34c-3p and SLC7A11. qRT-PCR (C) and western blotting (D) were performed to examine SLC7A11 mRNA and protein levels. (E, F) Flow cytometry was performed to detect the expression of CD86 and CD206. * P < 0.05, *** P < 0.001.
Figure 6. miR-34c-3p blocks NPC progression in vivo. miR-34c-3p mimic was injected into the xenograft model, and the tumor volume (A, B) and weight (C) were measured. (D) HE staining was performed to assess the pathological changes in tumor tissues. (E, F) Immunohistochemical staining was performed to detect CD86 and CD206 expression in tumor tissues. * P < 0.05, ** P < 0.01, *** P < 0.001.
Table 1. The primers of genes
Gene Sequences (5′–3′) iNOS (forward) TTCAGTATCACAACCTCAGCAAG iNOS (reverse) TGGACCTGCAAGTTAAAATCCC Arg-1 (forward) GTGGAAACTTGCATGGACAAC Arg-1 (reverse) AATCCTGGCACATCGGGAATC IL-10 (forward) GACTTTAAGGGTTACCTGGGTTG IL-10 (reverse) TCACATGCGCCTTGATGTCTG IL-6 (forward) ACTCACCTCTTCAGAACGAATTG IL-6 (reverse) CCATCTTTGGAAGGTTCAGGTTG TNF-α (forward) CCTCTCTCTAATCAGCCCTCTG TNF-α (reverse) GAGGACCTGGGAGTAGATGAG SLC7A11 (forward) TCTCCAAAGGAGGTTACCTGC SLC7A11 (reverse) AGACTCCCCTCAGTAAAGTGAC β-actin (forward) AGCGAGCATCCCCCAAAGTT β-actin (reverse) GGGCACGAAGGCTCATCATT -
[1] Xiang ZF, Hu DF, Xiong HC, et al. Benefit of chemotherapy in stage III nasopharyngeal carcinoma: analysis of the surveillance, epidemiology, and end results database. Oral Oncol, 2021; 117, 105284. doi: 10.1016/j.oraloncology.2021.105284 [2] Fan M, Liu DQ, Zhu GQ, et al. Comprehensive treatment of recurrent and metastatic nasopharyngeal carcinoma: advances and future directions. Precis Radiat Oncol, 2022; 6, 328−34. doi: 10.1002/pro6.1181 [3] Yan G, Feng Y, Wu MY, et al. Prognostic significance of MRI-based late-course tumor volume in locoregionally advanced nasopharyngeal carcinoma. Radiat Oncol, 2022; 17, 111. doi: 10.1186/s13014-022-02087-2 [4] Chen JJW, Lin YC, Yao PL, et al. Tumor-associated macrophages: the double-edged sword in cancer progression. J Clin Oncol 2025; 23, 953–64. [5] Zhou JW, Tang ZW, Gao SY, et al. Tumor-associated macrophages: recent insights and therapies. Front Oncol, 2020; 10, 188. doi: 10.3389/fonc.2020.00188 [6] Bouchery T, Volpe B, Doolan R, et al. β‐Glucan receptors on IL‐4 activated macrophages are required for hookworm larvae recognition and trapping. Immunol Cell Biol, 2022; 100, 223−34. doi: 10.1111/imcb.12536 [7] Zhang B, Miao TY, Shen X, et al. EB virus-induced ATR activation accelerates nasopharyngeal carcinoma growth via M2-type macrophages polarization. Cell Death Dis, 2020; 11, 742. doi: 10.1038/s41419-020-02925-9 [8] Ooft ML, Van Ipenburg JA, Sanders ME, et al. Prognostic role of tumour-associated macrophages and regulatory T cells in EBV-positive and EBV-negative nasopharyngeal carcinoma. J Clin Pathol, 2018; 71, 267−74. doi: 10.1136/jclinpath-2017-204664 [9] Chatterjee B, Saha P, Bose S, et al. MicroRNAs: as critical regulators of tumor- associated macrophages. Int J Mol Sci, 2020; 21, 7117. doi: 10.3390/ijms21197117 [10] Ma DH, Zhang Y, Chen GH, et al. miR-148a affects polarization of THP-1-derived macrophages and reduces recruitment of tumor-associated macrophages via targeting SIRPα. Cancer Manage Res, 2020; 12, 8067−77. doi: 10.2147/CMAR.S238317 [11] Zhang L, Liao Y, Tang LL. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res, 2019; 38, 53. doi: 10.1186/s13046-019-1059-5 [12] Zhang ZW, Sun CG, Li CC, et al. Upregulated MELK leads to doxorubicin chemoresistance and M2 macrophage polarization via the miR-34a/JAK2/STAT3 pathway in uterine leiomyosarcoma. Front Oncol, 2020; 10, 453. doi: 10.3389/fonc.2020.00453 [13] Pan Y, Hui XY, Hoo RLC, et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J Clin Invest, 2019; 129, 834−49. doi: 10.1172/JCI123069 [14] Duan C, Liu HR, Yang XQ, et al. Sirtuin1 inhibits calcium oxalate crystal-induced kidney injury by regulating TLR4 signaling and macrophage-mediated inflammatory activation. Cell Signal, 2023; 112, 110887. doi: 10.1016/j.cellsig.2023.110887 [15] Wei N, Lu T, Yang LB, et al. Lipoxin A4 protects primary spinal cord neurons from Erastin-nduced ferroptosis by activating the Akt/Nrf2/HO-1 signaling pathway. FEBS Open Bio, 2021; 11, 2118−26. doi: 10.1002/2211-5463.13203 [16] Chen D, Yu X. Long noncoding RNA TSLNC8 suppresses cell proliferation and metastasis and promotes cell apoptosis in human glioma. Mol Med Rep, 2018; 18, 5536−44. [17] Liu XY, Feng J, Tang LL, et al. The regulation and function of miR-21-FOXO3a-miR-34b/c signaling in breast cancer. Int J Mol Sci, 2015; 16, 3148−62. doi: 10.3390/ijms16023148 [18] Kim JS, Kim EJ, Lee S, et al. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp Mol Med, 2019; 51, 1−10. [19] He WF, Zhou XQ, Mao YN, et al. CircCRIM1 promotes nasopharyngeal carcinoma progression via the miR-34c-5p/FOSL1 axis. Eur J Med Res, 2022; 27, 59. doi: 10.1186/s40001-022-00667-2 [20] Huang W, Song W, Jiang YF, et al. c-Myc-induced circ-NOTCH1 promotes aggressive phenotypes of nasopharyngeal carcinoma cells by regulating the miR-34c-5p/c-Myc axis. Cell Biol Int, 2021; 45, 1436−47. doi: 10.1002/cbin.11582 [21] Luo ZH, Zhang LY, Li Z, et al. An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma. BMC Med Genomics, 2012; 5, 3. doi: 10.1186/1755-8794-5-3 [22] Wu ZD, Wu YP, Tian Y, et al. Differential effects of miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and invasion of glioma cells. Oncol Lett, 2013; 6, 1447−52. doi: 10.3892/ol.2013.1579 [23] Wu J, Li WZ, Huang ML, et al. Regulation of cancerous progression and epithelial-mesenchymal transition by miR-34c-3p via modulation of MAP3K2 signaling in triple-negative breast cancer cells. Biochem Biophys Res Commun, 2017; 483, 10−6. doi: 10.1016/j.bbrc.2017.01.023 [24] Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol, 2018; 233, 6425−40. [25] Liu WX, Chen GC, Zhang CY, et al. Prognostic significance of tumor-infiltrating lymphocytes and macrophages in nasopharyngeal carcinoma: a systematic review and meta-analysis. Eur Arch Otorhinolaryngol, 2022; 279, 25−35. doi: 10.1007/s00405-021-06879-2 [26] Liu Q, Yang T, Zhang Y, et al. ZIC2 induces pro-tumor macrophage polarization in nasopharyngeal carcinoma by activating the JUNB/MCSF axis. Cell Death Dis, 2023; 14, 455. doi: 10.1038/s41419-023-05983-x [27] Zhao SL, Mi YS, Guan BJ, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol, 2020; 13, 156. doi: 10.1186/s13045-020-00991-2 [28] Qiu SK, Xie L, Lu C, et al. Gastric cancer-derived exosomal miR-519a-3p promotes liver metastasis by inducing intrahepatic M2-like macrophage-mediated angiogenesis. J Exp Clin Cancer Res, 2022; 41, 296. doi: 10.1186/s13046-022-02499-8 [29] Wang PP, Wang HH, Huang QQ, et al. Exosomes from M1-polarized macrophages enhance paclitaxel antitumor activity by activating macrophages-mediated inflammation. Theranostics, 2019; 9, 1714−27. doi: 10.7150/thno.30716 [30] Xu MH, Zhou CH, Weng JL, et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J Exp Clin Cancer Res, 2022; 41, 253. doi: 10.1186/s13046-022-02458-3 [31] Huang H, Liu J, Wu HY, et al. Ferroptosis-associated gene SLC7A11 is upregulated in NSCLC and correlated with patient’s poor prognosis: an integrated bioinformatics analysis. Pteridines, 2021; 32, 106−16. doi: 10.1515/pteridines-2020-0034 [32] Dai EY, Han L, Liu J, et al. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy, 2020; 16, 2069−83. doi: 10.1080/15548627.2020.1714209 -




 下载:
下载:



 Quick Links
Quick Links