-
Proteus mirabilis is abundant in soil and water, and although it is part of the normal human intestinal flora (along with Klebsiella species and Escherichia coli), it is known to cause serious infections in humans, with a fatality rate of 20%–50%[1]. Proteus mirabilis is intrinsically resistant to tetracycline, tigecycline, and colistin. The widespread and irregular use of antimicrobial agents, P. mirabilis antimicrobial resistance has mirabilis. They acquire antimicrobial resistance (AMR) by capturing plasmids, transposons, or other mobile elements harboring antimicrobial resistance genes. This mechanism allows the rapid selection and transmission of numerous AMR genes in clinical, veterinary, food production, transportation, and environmental settings.
New Delhi metallo-β-lactamase, abbreviated as NDM, is an enzyme that can break β-lactam ring of lactam antimicrobial agents. This enables the inactivation of the antimicrobial and mediates resistance to nearly all the types of β-lactam antimicrobial agents, such as penicillin, cephalosporins and carbapenems, except for aztreonam. In recent years, reports of carbapenem-resistant bacteria in livestock, wild animals, food products, and the environment have increased[2]. However, only a few studies have focused on carbapenem-resistant P. mirabilis in humans.
The gene cfr was first identified in a Staphylococcus sciuri strain in 2,000 in the nasal cavity of calves suffering from respiratory diseases[3]. Functional research has revealed that cfr can mediate resistance to five classes of antimicrobial agents: chloramphenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A compounds through methylation of 23S rRNA, and it is most frequently located on plasmids[3]. Although the cfr gene was first discovered in Staphylococcus sciuri, it has been detected in gram-positive bacteria (Enterococcus[4], Streptococcus[5], Bacillus[6], etc.) from cattle, swine, and horses, and gram-negative bacteria (E. coli, Vibrio, Salmonella[7-8], etc.) from fish, chickens, and pigs.
In this study, we report the first instance of the coexistence of a novel blaNDM-1 plasmid and a novel chromosomal element carrying the cfr gene in a multidrug-resistant P. mirabilis YN_62 strain recovered from a human in Yunnan. This strain was susceptible to aztreonam and amikacin. This is the first report of such a combination in a human clinical isolate.
P. mirabilis strain YN_62 was isolated from a urine sample of a male patient with respiratory failure at Yunnan Baoshan Hospital in Yunnan Province, China, on November 18, 2022. The species was identified using BD Phoenix™ automated system (BD Diagnostic Systems, Sparks, MD, USA), and the Minimum inhibitory concentration (MIC) was tested by the micro broth dilution method according to Clinical and Laboratory Standards Institute guidelines. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega) and a bacterial genomic DNA extraction kit. The whole genome was sequenced using a PacBio RS II platform combined with an Illumina HiSeq platform.
The genome sequence was further polished with Illumina data using Pilon v1.22 (https://github.com/ broadinstitute/pilon/). Antimicrobial resistance genes were identified using ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster:ca/). Insertion sequences and transposons were identified using ISfinder (https://www-is.biotoul.fr/) and ICEfinder (http://bioinfo-mml.sjtu.edu.cn/ICEfinder/), respectively. A conjugation experiment was performed using the Escherichia coli J53 strain with sodium azide resistance as the recipient according to an established method[9].
Antimicrobial susceptibility testing showed that strain YN_62 was resistant to ampicillin, ampicillin-sulbactam, cefotaxime, cefoxitin, ceftazidime, ertapenem, imipenem, meropenem, chloramphenicol, colistin, streptomycin, tetracycline, tigecycline, trimethoprim-sulfamethoxazole, azithromycin, nalidixic acid, nitrofurantoin, and ciprofloxacin, and susceptible to amikacin and aztreonam (Table 1).
Table 1. Drug susceptibility patterns and antibiotic resistance genes of strain YN_62
Antimicrobial agent MIC (µg/mL) Int. Phenotype Resistance gene Ampicillin > 32 R Beta-lactam resistance blaCTX-M-65 Ampicillin-Sulbactam > 32/16 Cefotaxime > 16 Cefoxitin > 64 Ceftazidime > 16 Ceftazidime avibactam > 8/4 Chloramphenicol > 32 R Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A resistance cfr floR cat lnu(F) msr(E) Streptomycin > 32 R Aminoglycoside resistance aph(3')-Ia bleO aadA2b aadA1 Tetracycline > 16 R Tetracycline resistance tet(J) Tigecycline 8 Trimethoprim-
Sulfamethoxazole> 8/152 R Sulphonamide resistance dfrA1 sul1 sul2 Azithromycin > 64 R Macrolide resistance mph(E) msr(E) ere(A) Nalidixic Acid > 32 R Quinolone resistance qnrA1 Nitrofurantoin 64 Ciprofloxacin > 2 Ertapenem > 8 R Carbapenems blaNDM-1 Imipenem > 8 Meropenem > 8 Amikacin 8 S − − Aztreonam 0.5 S Colistin > 8 R Polymyxins − Note. Int., interpreted MIC results; R, resistant; S, susceptible. Genomic analysis revealed that strain YN_62 harbored a chromosome and circular plasmid named pYN 62-blaNDM-1. The chromosome was 4100124 bp in length with a GC content of 38.91 %. Antimicrobial resistance genes screening identified that the chromosome harbored the β-lactamase genes blaCTX-M-65 as well as other genes conferring resistance to aminoglycoside (aph(3')-Ia, aac(3)-IV, aph(4)-Ia, aadA2b, aadA1), quinolones (qnrA1), tetracyclines (tetC, tetJ), chloramphenicol (floR, cat), lincosamides and macrolide resistance (lnuF msrE, mphE), fosfomycin (fosA3) and cfr which mediated resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins and streptogramin A compounds.
Mutations in quinolone resistance-determining regions (QRDRs) of gyrA and gyrB were analyzed. Substitutions were observed at GyrA (Ser 83 to Ile) (Val 444 to Met) and GyrB (Glu 466 to Asp), as determined by alignment with the reference strain, P. mirabilis HI4320, in the GenBank database using NCBI nucleotide blast analysis. The Ser-83 to Ile mutation in GyrA is an important mechanism for mediating resistance to quinolone drugs. Importantly, all strains harboring gyrA mutations were resistant to quinolones.
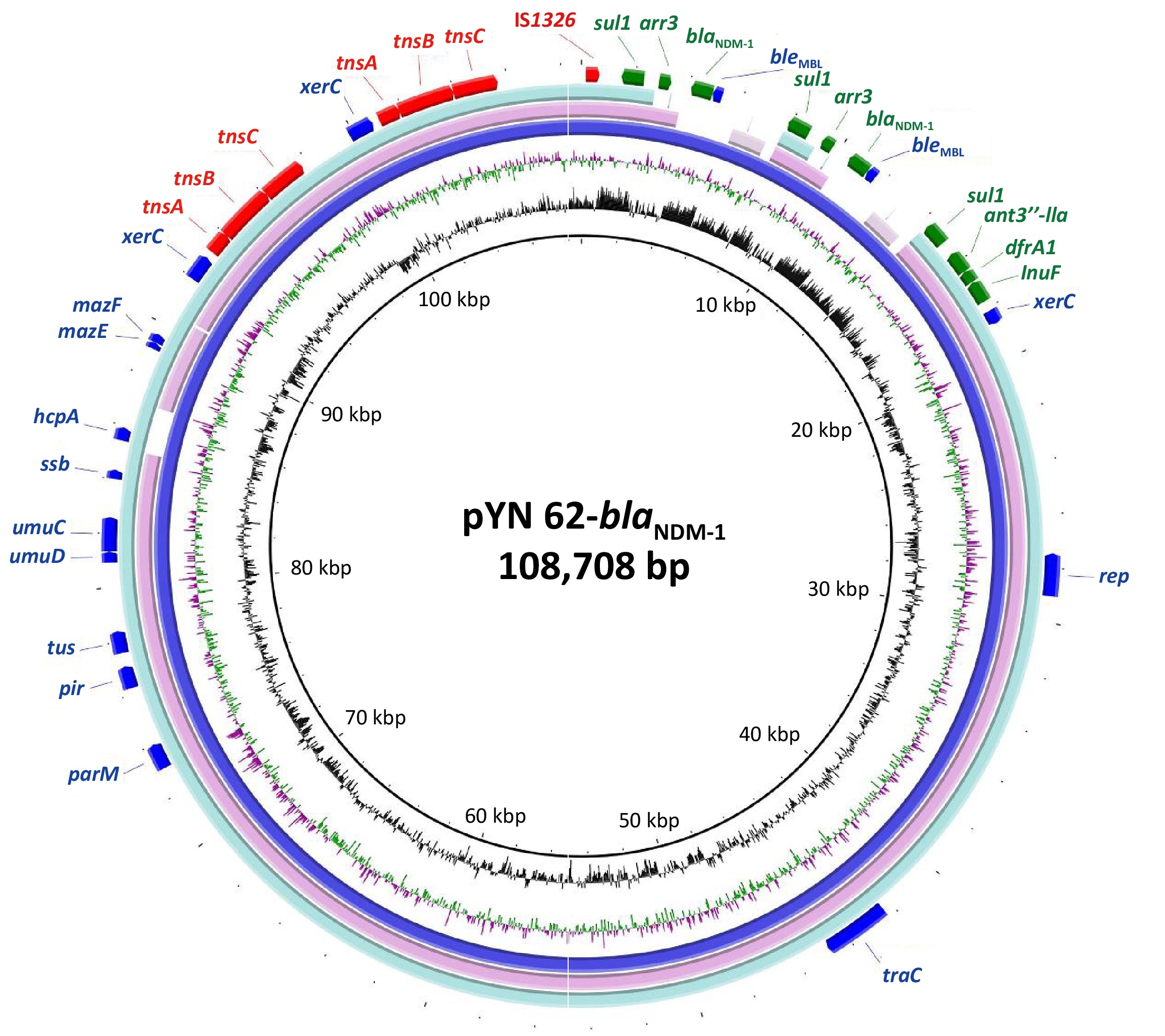
Plasmid pYN 62-blaNDM-1 was a closed circular DNA, 108.7 kb in length, with 40.7% mean G+C content and an unknown replication type. It carried three drug resistance gene clusters, one gene cluster (sul1-ant3′′-lla-dfrA1-InuF), and two duplicate copies of the gene cluster blaNDM-1 (sul1-arr3-blaNDM-1), which mediated resistance to quinolones, sulphonamide, chloramphenicol, aminoglycoside, and nearly all the β-lactams drugs including carbapenems. The backbone of the plasmid pYN 62-blaNDM-1 was closely related to the plasmids deposited in GenBank. One of these was strain G11, a 152.8-kb plasmid carrying sul1 gene from Proteus terrae subsp. cibarius (GenBank accession number: CP047288.1), showing > 99% nucleotide identity over > 68.5% of the backbone regions. Another was pW17-3-a, a 102.8-kb plasmid with a β-lactamase extended-spectrum beta-lactamase (ESBL) antimicrobial resistance gene from the Moellerella wisconsensis strain, with > 99% nucleotide identity over > 83.2% of the backbone regions, accession number CP093261.1 (Figure 1).
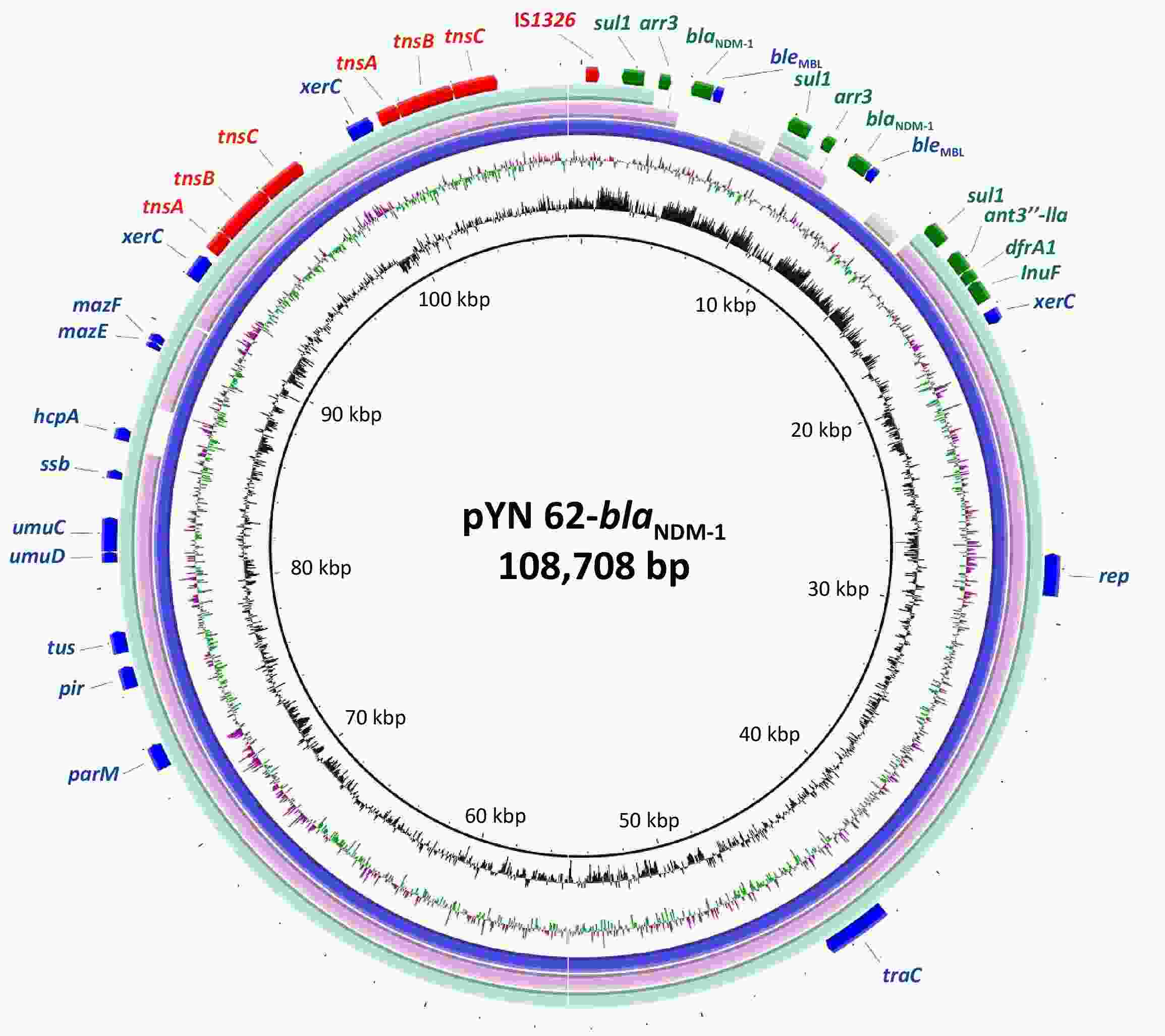
Figure 1. The circular map of the NDM-positive plasmid pYN 62-blaNDM-1 compared to other reported similar plasmids p152 (Proteus terrae subsp. cibarius strain G11; GenBank: CP047288.1), p3804_NDM (Enterobacter hormaechei strain 3804; GenBank: CP064660.1. in the NCBI database. The innermost circle provides a size scale, while the next innermost circle shows the GC content.
The transconjugants were selected on Mueller-Hinton agar plates supplemented with 200 mg/L sodium azide and 2 mg/L imipenem and confirmed by antimicrobial susceptibility testing and PCR to detect blaNDM-1 gene[9]. Unfortunately, positive Escherichia coli J53 transconjugants carrying the 62-blaNDM-1 plasmid were not obtained, indicating that the conjugation transfer efficiency of the 62-blaNDM-1 plasmid was relatively low.
The cfr gene is most frequently found in plasmids; however, it has also been described on rare occasions within integrative and conjugative elements (ICEs) located on chromosomes. To date, only two ICEs carrying the cfr gene have been identified: ICEPmiChnBCP11 (accession number 201) and ICEPvuChnBC22 (accession number 200). These ICEs have been found in P. mirabilis and P. vulgaris isolated from the fecal swabs of pigs with diarrhea in China[10]. To the best of our knowledge, the presence of the multi-resistance gene cfr in P. mirabilis isolated from humans is reported for the first time. This gene is encoded by a putative ICE within the chromosomal nucleotide sequence and is flanked by multiple copies of the IS26 element, such as IS26-bleO-IS26-cfr-IS26-btuB-IS5-blaCTX-M-65-IS26-aac(3)-IV-aph(3)-IV. Notably, any two copies of IS26 have the potential to mediate the mobilization of the cfr resistance gene.
BLAST analysis showed that some partial regions, including the upstream region IS26-bleO-IS26-cfr-IS26, have high homology with P. mirabilis PM14 isolated from humans (Zhejiang accession number GenBank: CP137085.1), STP3 isolated from pigs in Sichuan Province, China (accession number GenBank: CP051260.1), and the downstream region btuB-IS5-blaCTX-M-65-IS26-aac(3)-IV-aph(3)-IV-IS6-Tn3-IS4-IS4-sul2 -glmM-hdfR-floR-Tn3-IS3000-umuC-ints has high homology with P. mirabilis SNYG35 isolated from chicken in Sichuan Province, China, accession number GenBank: CP047589.1 (Figure 2).
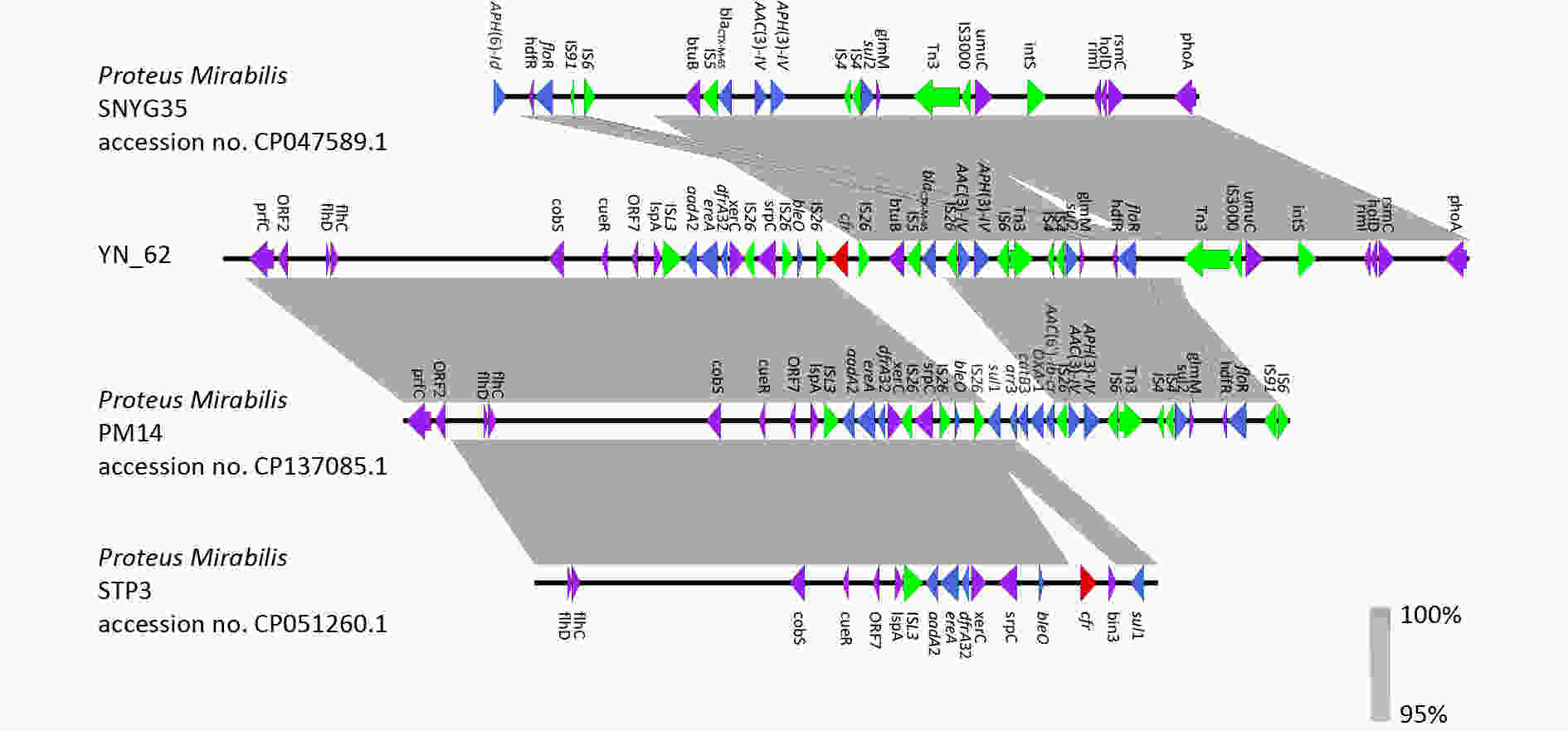
Figure 2. Comparative analysis of fragment carrying cfr in different strains. Strains information is listed on the left side. ORFs are indicated by arrows. Red arrows indicate the cfr gene. Blue arrows indicate the other antimicrobial resistance genes. Green arrows indicate the insertion sequences. Purple arrows indicate other genes. Identical regions are highlighted in gray (> 95% similarity).
The blaNDM-1 and cfr genes are clinically significant because of their roles in conferring resistance to critical antimicrobial agents. In this study, a strain harboring both genes was identified in urine samples from patients with urinary tract infections, representing a substantial threat to treatment outcomes and overall prognosis. The fact that blaNDM-1 is located on a plasmid and the cfr gene is adjacent to the IS26 element suggests that these resistance determinants may be mobile and capable of being disseminated to other bacterial strains, thereby increasing the risk of antimicrobial resistance. Therefore, there is an urgent requirement for stepped-up surveillance of blaNDM-1 and cfr resistance elements to curtail their transmission within communities, particularly within healthcare facilities.
doi: 10.3967/bes2024.146
Emergence of Multidrug-Resistant Proteus mirabilis Harboring both blaNDM-1 and cfr genes
-
-
Figure 1. The circular map of the NDM-positive plasmid pYN 62-blaNDM-1 compared to other reported similar plasmids p152 (Proteus terrae subsp. cibarius strain G11; GenBank: CP047288.1), p3804_NDM (Enterobacter hormaechei strain 3804; GenBank: CP064660.1. in the NCBI database. The innermost circle provides a size scale, while the next innermost circle shows the GC content.
Figure 2. Comparative analysis of fragment carrying cfr in different strains. Strains information is listed on the left side. ORFs are indicated by arrows. Red arrows indicate the cfr gene. Blue arrows indicate the other antimicrobial resistance genes. Green arrows indicate the insertion sequences. Purple arrows indicate other genes. Identical regions are highlighted in gray (> 95% similarity).
Table 1. Drug susceptibility patterns and antibiotic resistance genes of strain YN_62
Antimicrobial agent MIC (µg/mL) Int. Phenotype Resistance gene Ampicillin > 32 R Beta-lactam resistance blaCTX-M-65 Ampicillin-Sulbactam > 32/16 Cefotaxime > 16 Cefoxitin > 64 Ceftazidime > 16 Ceftazidime avibactam > 8/4 Chloramphenicol > 32 R Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A resistance cfr floR cat lnu(F) msr(E) Streptomycin > 32 R Aminoglycoside resistance aph(3')-Ia bleO aadA2b aadA1 Tetracycline > 16 R Tetracycline resistance tet(J) Tigecycline 8 Trimethoprim-
Sulfamethoxazole> 8/152 R Sulphonamide resistance dfrA1 sul1 sul2 Azithromycin > 64 R Macrolide resistance mph(E) msr(E) ere(A) Nalidixic Acid > 32 R Quinolone resistance qnrA1 Nitrofurantoin 64 Ciprofloxacin > 2 Ertapenem > 8 R Carbapenems blaNDM-1 Imipenem > 8 Meropenem > 8 Amikacin 8 S − − Aztreonam 0.5 S Colistin > 8 R Polymyxins − Note. Int., interpreted MIC results; R, resistant; S, susceptible. -
[1] Chen SL, Kang YT, Liang YH, et al. A core genome multilocus sequence typing scheme for Proteus mirabilis. Biomed Environ Sci, 2023; 36, 343−52. [2] Xie X, Zhang JL, Wang HN, et al. Whole genome sequence of a New Delhi metallo-β-lactamase 1-producing Proteus mirabilis isolate SNYG35 from broiler chicken in China. J Glob Antimicrob Resist, 2021; 24, 266−9. doi: 10.1016/j.jgar.2020.12.023 [3] Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother, 2000; 44, 2530−3. doi: 10.1128/AAC.44.9.2530-2533.2000 [4] Chen Q, Yin D, Li P, et al. First report Cfr and OptrA co-harboring linezolid-resistant Enterococcus faecalis in China. Infect Drug Resist, 2020; 13, 3919−22. doi: 10.2147/IDR.S270701 [5] Wang Y, Li D, Song L, et al. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob Agents Chemother, 2013; 57, 4061−3. doi: 10.1128/AAC.00713-13 [6] Dai L, Wu CM, Wang MG, et al. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob Agents Chemother, 2010; 54, 3953−5. doi: 10.1128/AAC.00169-10 [7] Liu M, Zhang WH, Hu YF, et al. Emergence of the cfr gene in Vibrio diabolicus of seafood origin. Antimicrob Agents Ch, 2022; 66, e01819−21. [8] Hara GL. Emergence of plasmid harbouring the cfr gene in porcine Salmonella. Int J Antimicrob Agents, 2023; 62, 106833. doi: 10.1016/j.ijantimicag.2023.106833 [9] Yuan M, Chen H, Zhu X, et al. pSY153-MDR, a p12969-DIM-related mega plasmid carrying blaIMP-45 and armA, from clinical Pseudomonas putida. Oncotarget, 2017; 8, 68439−47. doi: 10.18632/oncotarget.19496 [10] Lei CW, Chen YP, Kang ZZ, et al. Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, blaCTX-M-65, fosA3, and aac(6')-Ib-cr in Proteus mirabilis. Antimicrob Agents Chemother, 2018; 62, e00849−18. -




 下载:
下载:



 Quick Links
Quick Links