-
Autoimmune hepatitis (AIH) is a chronic inflammatory liver disease caused by a disorder in the immune system and can develop into liver fibrosis, cirrhosis, and eventually liver cancer until death[1]. AIH is prevalent in women and disease incidence is currently showing an increasing trend worldwide; however, its pathogenesis remains unclear[2-3]. Concanavalin A (Con A) injection is a typical method to induce AIH in experimental animals[4]. The generated responses are characterized by induction of infiltration of T cells, macrophages, neutrophils, and natural killer cells (NKT) in the liver tissue[5]. Here, we chose the Con A model to investigate the mechanisms underlying AIH and possible treatment options.
Macrophage polarization is essential to the development of AIH[6-7]. Several clinical studies have convincingly demonstrated that overexpression of M1 macrophages plays a key role in inflammatory responses and liver injury in patients with AIH[8]. Studies have revealed that when liver cells are exposed to external stimuli in patients with liver disease, large numbers of lymphocytes and macrophages rapidly accumulate and become activated, causing a series of immune responses. Under inflammatory conditions, macrophages polarize toward the M1 phenotype[9-10]. However, different from non-alcoholic fatty liver disease and chronic hepatitis B, patients with AIH have higher M1 cell counts. Besides, macrophage activation correlates with AIH activity and severity, but the exact mechanism is still under investigation[11].
C6orf120, a gene of unknown function that encodes an N-glycosylated protein, has high homology between humans and rats; therefore, disease development in humans can be successfully mimicked in rat models[12]. Previous studies by our group demonstrated that C6orf120 knockout is essential for immune regulation in the Con A-induced AIH model in rats. C6orf120 knockout promotes the apoptosis of CD4+ T cells, increases the frequency of Treg cells, and inhibits the activation of NKT cells and the secretion of inflammatory factors, among other functions[12-14]. However, the potential regulation of macrophage polarization by C6orf120 and the associated mechanism have not been thoroughly investigated. In this study, we constructed an AIH rat model to investigate in depth the immunomodulatory mechanism of C6orf120 on AIH. The C6orf120-knockout macrophage cell line (THP-1 cells) and the recombinant protein (rC6ORF120) were constructed for in vitro experiments.
-
Male wild-type (WT) Sprague Dawley rats (180–200 g, 6–8 weeks old) were supplied by the Laboratory Animal Center of Health Science Center, Peking University. Male knockout rats (C6orf120-/-; 180–200 g, 6–8 weeks old) were provided by Guangzhou Saiye Biotechnology Co., Ltd., using TALEN-mediated gene knockout technology. This technique can cause a partial base-shift mutation in the C6orf120 gene, making the transcriptional translation of the C6ORF120 protein unsuccessful[14]. To verify the reliability of the knockout, we obtained tissues from the tail from WT and C6orf120-/- rats for gene identification. Both WT and C6orf120-/- rats were kept in a pathogen-free environment (3 rats/cage, 21 ± 2 °C, 50% relative humidity, 12/12 h daytime/nighttime artificial light cycle, and water and food at all times) at the Laboratory Animal Center of Health Science Center, Peking University.
To establish an experimental AIH model in rats induced by Con A, WT and C6orf120-/- rats aged 6–8 weeks were randomly divided into two groups; the control group was administered saline via tail-vein injection and the experimental group was administered Con A dissolved in saline (16 mg/kg) (Sigma, Aldrich, USA). After 24 h, the rats were euthanized, and the liver, spleen, and inferior vena cava blood were collected for further studies. All animal experiments followed the NIH Guide for the Care and Use of Laboratory Animals[15].
-
PCR and gene sequencing were used to identify the rat genotypes. DNA was extracted from the rat tails using an animal tissue DNA extraction kit (Invitrogen, MA, USA). Subsequently, the DNA was amplified using PCR. The specific amplification method was as follows: initial 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; the termination step was 72 °C for 1 min. The specific primer sequences for C6orf120 are Forward: 5′-AGCACCTCCGGTCAAGTCTGTCAC-3′ and Reverse: 3′-GTCGGACACATACAGGTCCGCA-5′. The final amplified DNA sequences of WT and C6orf120-/- rats were compared to identify specific and complete knockouts of the C6orf120 gene.
-
Serum from the peripheral veins of patients with AIH (n = 15) and healthy volunteers (n = 15) was collected at Beijing Ditan Hospital. All procedures were carried out under the supervision of the Ethics Committee of Beijing Ditan Hospital, Capital Medical University.
-
A human C6ORF120 ELISA kit purchased from Mlbio (Shanghai, China) was used to detect C6ORF120 protein levels in the serum of patients with AIH and healthy individuals. All operations were performed according to the instruction manual.
-
Whole blood samples from the inferior vena cava were obtained and centrifuged at 3,000 rpm at 4 °C for 10 min to obtain serum samples. ALT and AST levels in the rat serum were measured using a HITACHI instrument according to the method standards provided by the manufacturer.
-
After euthanizing the rats, the liver tissue was fixed in 4% paraformaldehyde. After dehydration and paraffin embedding, the liver tissue was cut into 4-µm thick sections to stain for H&E. Photographs were acquired under a microscope (Zeiss AG, Germany).
-
The degree of liver damage was assessed using the Ishak score[16]. Liver pathology is mainly assessed based on three major aspects: necrotic area (0 for no necrosis, 1 for < 10% of hepatic parenchyma necrosis, 2 for 10%–25%, and 3 for > 25%), lobular inflammation (0 for no inflammation, 1 for < 10% of hepatic parenchyma inflammation, 2 for 10%–50%, and 3 for > 50%), and portal inflammation (0 for no inflammation, 1 for < 1/3 of the area, 2 for 1/3–1/2 of the area, and 3 for > 1/2 of the area), with the overall score being the Tissue Inflammation Score. The pathology score for the extent of liver injury was assessed by two independent authors (Hui Liu and Yingying Lin) who are well-trained and were blinded to group allocation.
-
THP-1 cells were obtained from Wuhan Procell Technology Co. and cultured in Dulbecco's Modified Eagle Medium (DMEM), containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were grown in a 37 °C incubator with 5% CO2. Phorbol 12-myristate 13-acetate (PMA) (100 ng/mL), lipopolysaccharide (LPS) (100 ng/mL), gamma interferon γ (INF-γ) (20 ng/mL), interleukin 13 (IL13) (20 ng/mL), interleukin 4 (IL4) (20 ng/mL), and rC6ORF120 (Cusabio, Wuhan, China) were used to stimulate THP-1 cells.
-
THP-1 cells were equally inoculated in 12-well plates, and PMA (100 ng/mL) was added to stabilize the cells to adhere to the wall and become mature M0 macrophages. The siRNA was synthesized by Shanghai GenePharma Technology Co. The sequence of the RNA targeting the C6orf120 gene (siC6orf120) is 5′-GCGAGUUCGAGAUGAAGGUTT-3′, and the non-specific RNA sequence is 5′-UUCUCCGAACGUGUCACGUTT-3′. The siRNA expression vectors were transfected into THP-1 cells using Lipofectamine 3000 (Invitrogen, MA, USA).
-
Tissues obtained from rats were lysed using RIPA lysis buffer (Gene-protein link, Beijing, China). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate proteins extracted from the liver or spleen tissues. The proteins were then transferred wet onto polyvinylidene fluoride membranes (PVDF membranes). After blocking with 5% skimmed milk, the following primary antibodies were incubated with membranes overnight at 4 °C: C6ORF120 (bs-9354R; Bioss, Beijing, China; 1:500 dilution), CD206 (18704-1-AP; Proteintech Group, Chicago, IL, USA; 1:1,000 dilution), and CD86 (bs-1035R; Bioss; 1:500 dilution). The strips were then incubated with a secondary immunoglobulin antibody on a shaker at room temperature for 1 h. Finally, the target strips were detected using chemiluminescence.
-
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and tested for RNA concentration and purity using a spectrophotometer. The RNA was reverse-transcribed into cDNA using the PrimeScriptTM RT reagent Kit (RR037A; TaKaRa, Shiga, Japan) according to the manufacturer's instructions, and the RNA was extracted using the Power SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific). RT-qPCR was performed using a standard protocol. GAPDH was used as the reference and all data were analyzed according to 2-ΔΔCT. All primer sequences are listed in Table 1.
Table 1. Primer sequence for RT-qPCR
Gene (ID) Species Forward (5‘-3’) Reverse (5‘-3’) GAPDH Rat GGCATCGTGGAAGGGCTCAT CGTCGGGTCTTGTAGTAGGGA TNF-α Rat GCGATGTGGAACTGGCAGAGG GAGAAGAGTAAGGACGAGCACCG IL-1β Rat ATCTCACAGCAGCATCTCGACAAG CCTACTACTGCTGGACGATCACAC IL-6 Rat TTCCAGCCAGTTGCCTTCTT GTGAAGTGTTCAGCCTCCGAA ARG1 Rat CCAAGCCAAAGCCCATAGAGAT ACCAGGCCAGCTTTCCTTAAT IL-10 Rat GAAGGACCAGCTGGACAACA GGGGCATCACTTCTACCAGG GAPDH Human AGAAGGCTGGGGCTCATTTG AGGGGCCATCCACAGTCTTC CCL1 Human CTCATTTGCGGAGCAAGAGAT GCCTCTGAACCCATCCAACTG IL6 Human TGGCAGAAAACAACCTGAACC GGCTTGTTCCTCACTACTCTCA IL10 Human GGCATCTACAAAGCCATGAGTG TTTCTCAAGGGGCTGGGTCA CD206 Human GGGACGTGGCTGTGGATAAA TCCAAAACCCAGAAGACGCA TNF-α Human TGCACTTTGGAGTGATCGGC ACTCGGGGTTCGAGAAGATG CD80 Human TTGGTGCTGGCTGGTCTTTC TGCCAGTAGATGCGAGTTTGT -
When liver tissue was obtained from rats in vivo, phosphate-buffered saline-BSA-EDTA buffer (PBEB) was injected via the hepatic portal vein to remove intrahepatic red blood cells. The hepatocyte suspension was then passed through a 40-µm cell filter (BD, NJ, USA). The filtered cell suspension was resuspended in 15 mL of PBEB and centrifuged at 50 g, 4 °C for 3 min to remove the hepatic parenchymal cells. The supernatant was collected and centrifuged at 1,200 rpm, 4 °C for 5 min. The cells were resuspended in 40% Percoll and slowly added to the upper layer containing 4 mL of 80% Percoll and centrifuged at 450 g (acceleration 9, brake 1), 4 °C for 20 min. The intermediate cell layer was collected to prepare a cell suspension for flow analysis.
After obtaining the spleen tissue, a spleen cell suspension was prepared in PBEB to which 4 mL of erythrocyte lysis solution was added, mixed, and allowed to stand for 10 min to fully lyse the erythrocytes. The lysis process was terminated by adding 2 mL of pre-cooled PBEB buffer. Then the mixture was centrifuged at 1,200 rpm, 4 °C was performed for 5 min and the lower cell layer was obtained, dissolved in PBEB, and filtered for flow cytometric analysis.
-
The obtained liver and spleen single-cell suspensions were stained with anti-rat CD45 APC cy7 (Biolegend, CA, USA), anti-rat CD86 FITC (Biolegend), and anti-rat CD163 PE (Bio-Rad, CA, USA) antibodies at 4 °C for 30 min. THP-1 cells were stained with anti-human CD80 APC (eBioscience, CA, USA) and anti-human CD206 FITC (eBioscience) antibodies. To determine the fluorescence level of intracellular CD68, the cell membrane was disrupted in the presence of intracellular permeabilization buffer and stained with anti-rat CD68 APC (Miltenyi Biotec, North Rhine-Westphalia, Germany) and anti-human CD86 PEcy7 (eBioscience) antibody for 30 min at room temperature. Cell counting was performed using a BD FACS Canto II flow cytometer (BD Biosciences) and imaging data were analyzed using the FlowJo software (FlowJo, LLC, Ashland, OR).
-
Quantitative data are shown as the mean ± standard deviation and statistically processed using GraphPad Prism 9.0 (San Diego, CA, USA). The t-test, one-way ANOVA, two-way ANOVA, and Kruskal–Wallis test were used for the majority of the data. Statistical significance was considered at P < 0.05.
-
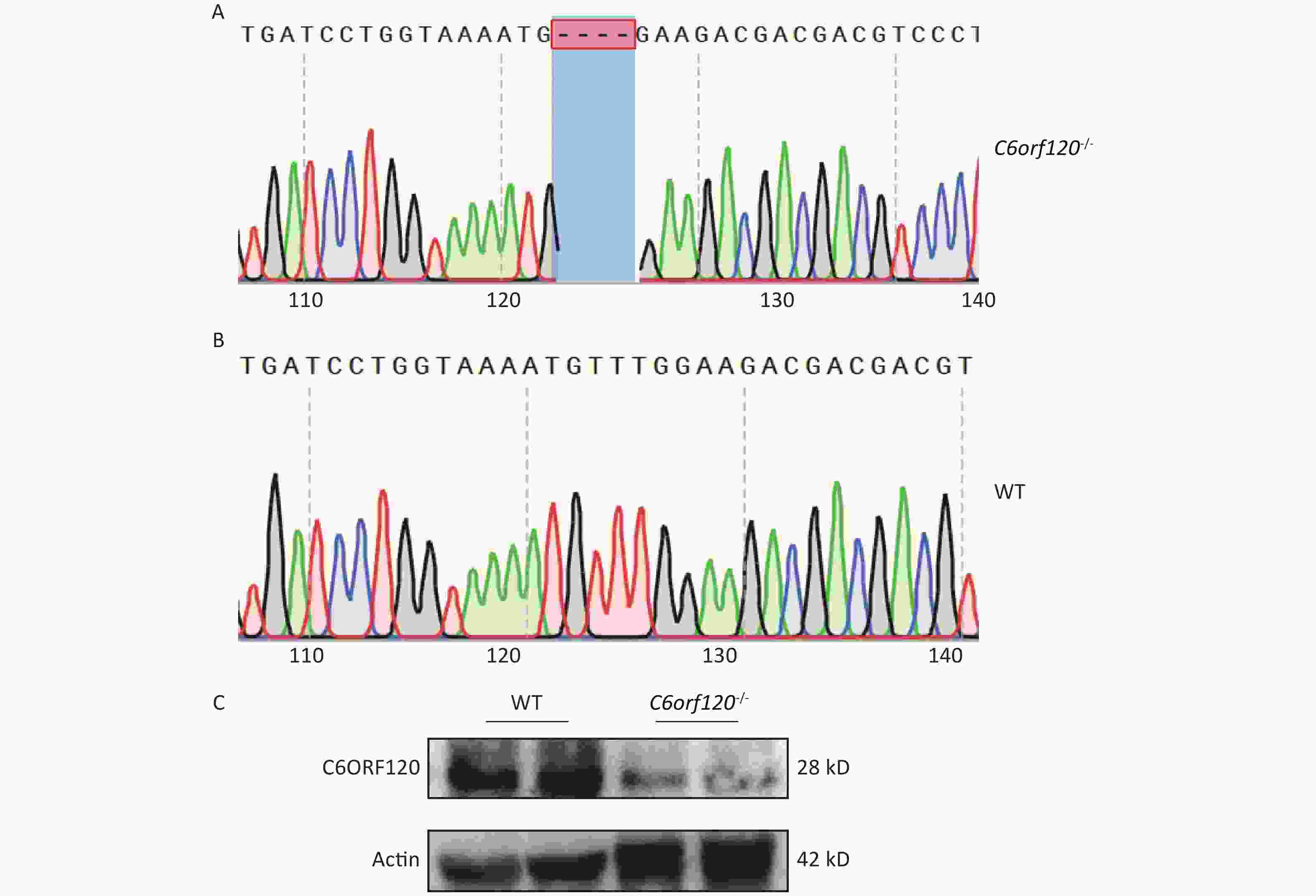
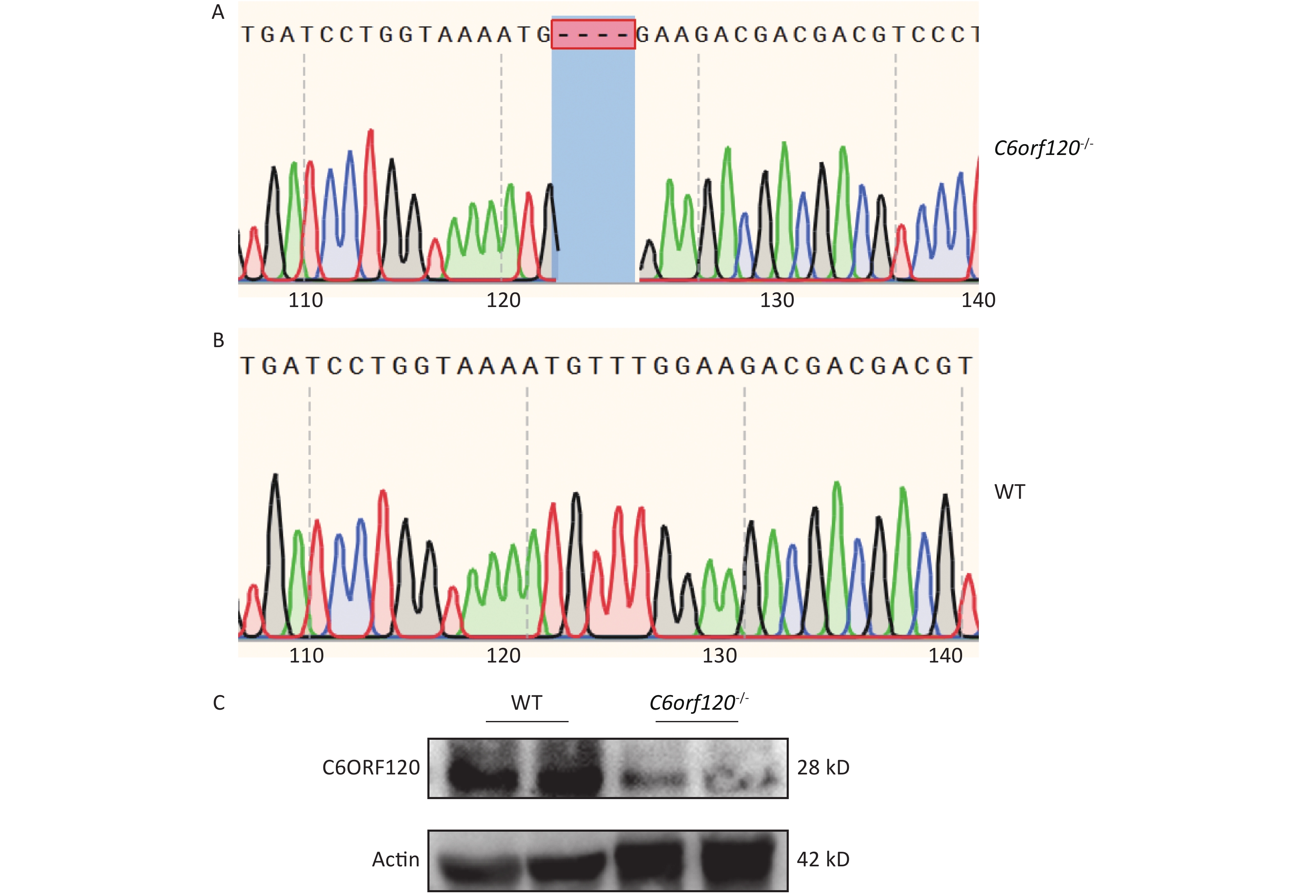
Our group used DNA sequencing and western blotting techniques to identify the rat genotypes. Figure 1A and B show the DNA sequences of WT and C6orf120-/- rats, respectively. C6orf120-/- rats had a deficiency in four bases (TTTG) compared with WT rats, resulting in unsuccessful expression of the C6ORF120 protein. The western blotting results showed that the expression of the C6ORF120 protein decreased sharply in the liver tissue of C6orf120-/- rats (Figure 1C).
-
In liver tissue from WT rats, we found that C6ORF120 protein expression increased significantly following Con A induction (Figure 2A). Furthermore, we found that the level of C6ORF120 in patients with AIH was higher than that in healthy volunteers (Figure 2A). To explore the role and mechanism associated with the functionally unknown gene C6orf120 in AIH, we selected WT and C6orf120-/- rats to mimic the presence and absence, respectively, of C6orf120 in humans. Interestingly, we found that C6orf120 knockout appeared to protect rats from Con A-induced AIH to some extent. C6orf120-/- rats exhibited lower levels of ALT (P < 0.05) and AST (P < 0.0001) than the WT rats in the Con A-induced AIH model (Figure 2D, E). In addition, histopathological analysis of the liver showed that WT rats had more extensive liver tissue damage, a more disorganized liver plate structure, and more pronounced inflammatory cell infiltration (Figure 2B, C). Taken together, these results suggest that C6orf120-/- rats withstand less damage in Con A-induced AIH.
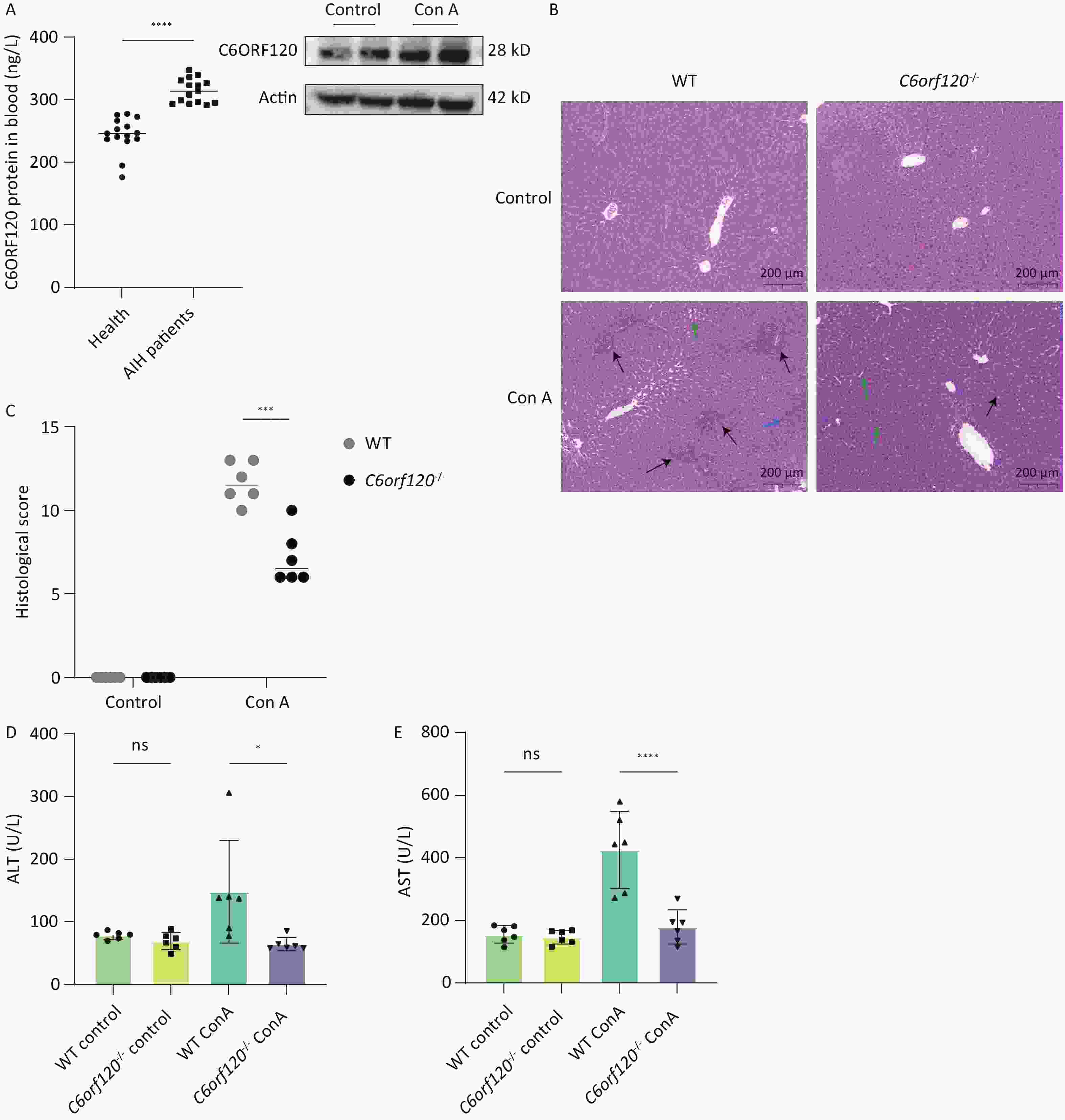
Figure 2. Assessment of liver injury. (A) Western blotting analyzing C6ORF120 protein in the liver tissue in the presence or absence of Con A stimulation; ELISA analyzing C6ORF120 protein level in healthy volunteers and patients with AIH. (B) H & E staining of liver sections (black arrows indicate inflammatory cell infiltration; green arrows, fatty degeneration; and blue arrows, necrosis). (C) Histopathological score of the liver. (D, E) Serum ALT and AST levels (n = 6). *P < 0.05; ***P < 0.001; and ****P < 0.0001. WT, wild-type; ns, no statistical significance.
-
Macrophages are highly plastic, and it is now generally recognized that they can phenotypically change to M1 macrophages, which are involved in the pro-inflammatory response against danger signals in the body. However, they also have the potential to differentiate into anti-inflammatory M2 macrophages, which participate in tissue repair, structural reconstruction, and inhibition of inflammation[17]. The development of AIH is closely associated with macrophage activation[11]. To investigate whether C6orf120-/- affects autoimmune liver injury by influencing macrophage activation, we analyzed the frequency of CD68+CD86+M1 and CD68+CD163+M2 macrophages in the rat liver and spleen after Con A stimulation using flow cytometry. The results showed that the number of CD68+CD86+M1 macrophages was significantly lower in both the liver (P < 0.0001) and spleen (P < 0.01) than in the tissues of WT rats (Figure 3A–C). At the same time, a significant increase was observed in the number of CD68+CD163+M2 macrophages in the spleen of C6orf120-/- rats (P < 0.0001), but no statistically significant difference in the liver, although the frequency of CD68+CD163+M2 macrophages increased (Figure 3D–F).
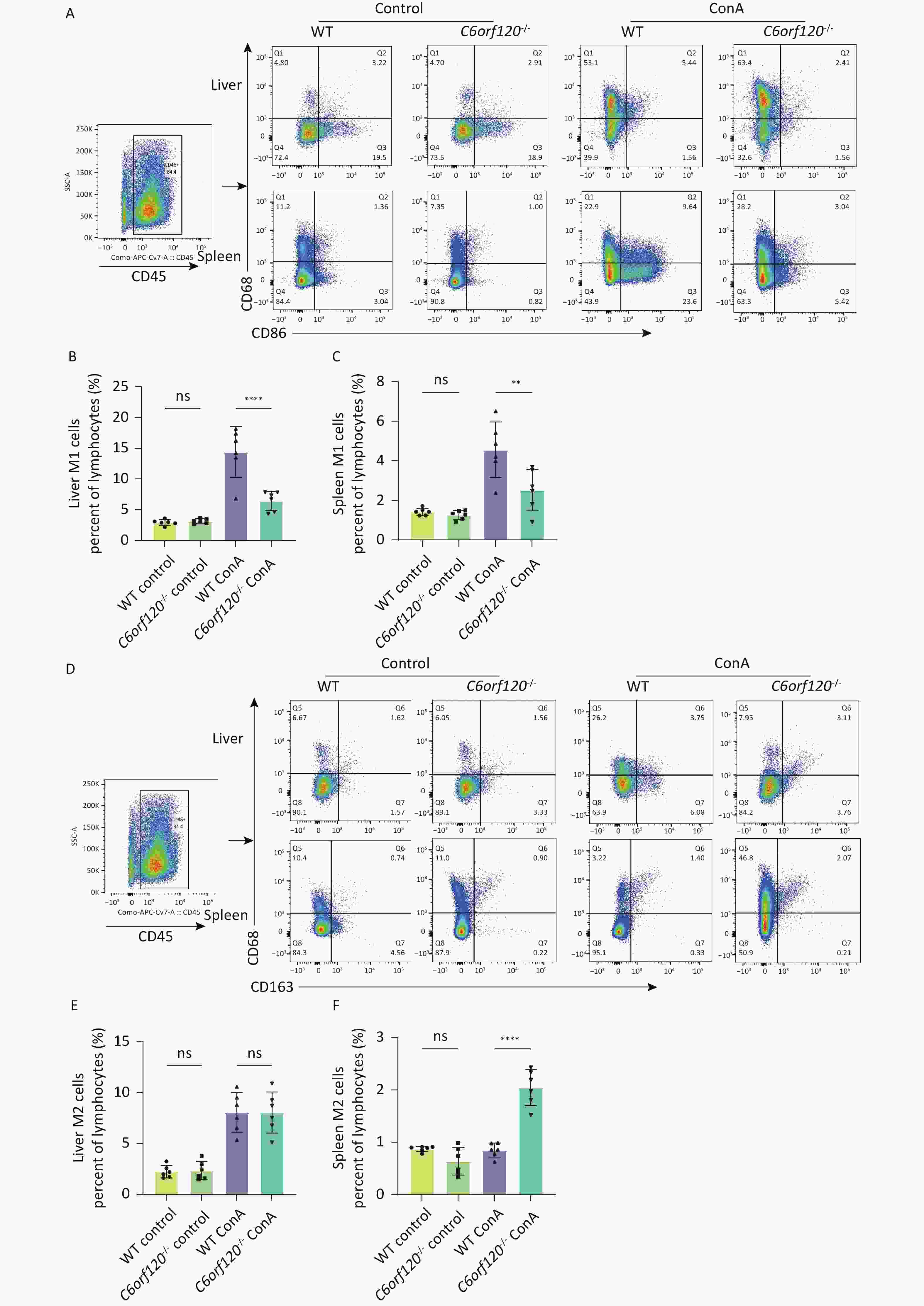
Figure 3. Macrophage subsets in vivo. (A) Representative image of the gating strategy of CD68+CD86+M1 macrophages in liver and spleen lymphocytes. Frequency of M1 macrophages in the liver (B) and spleen (C). (D) Representative images of the gating strategy of CD68+CD163+M2 macrophages in liver and spleen lymphocytes. Frequency of M2 macrophages in the liver (E) and spleen (F) (n = 6). **P < 0.01; ****P < 0.0001; WT, wild-type; ns, no statistical significance.
-
CD86 is a surface marker for M1 macrophages, while CD206 is a marker for M2 macrophages. To verify the reliability of the results in several respects, the expression of CD86 and CD206 proteins was examined using western blotting. The results showed that deletion of C6orf120 downregulated the expression of the CD86 protein in the liver (P < 0.001) (Figure 4A, B) and spleen tissues (P < 0.01) (Figure 4A, C), and, to some extent, upregulated the protein expression of CD206 in the spleen under Con A induction (Figure 4D–E).
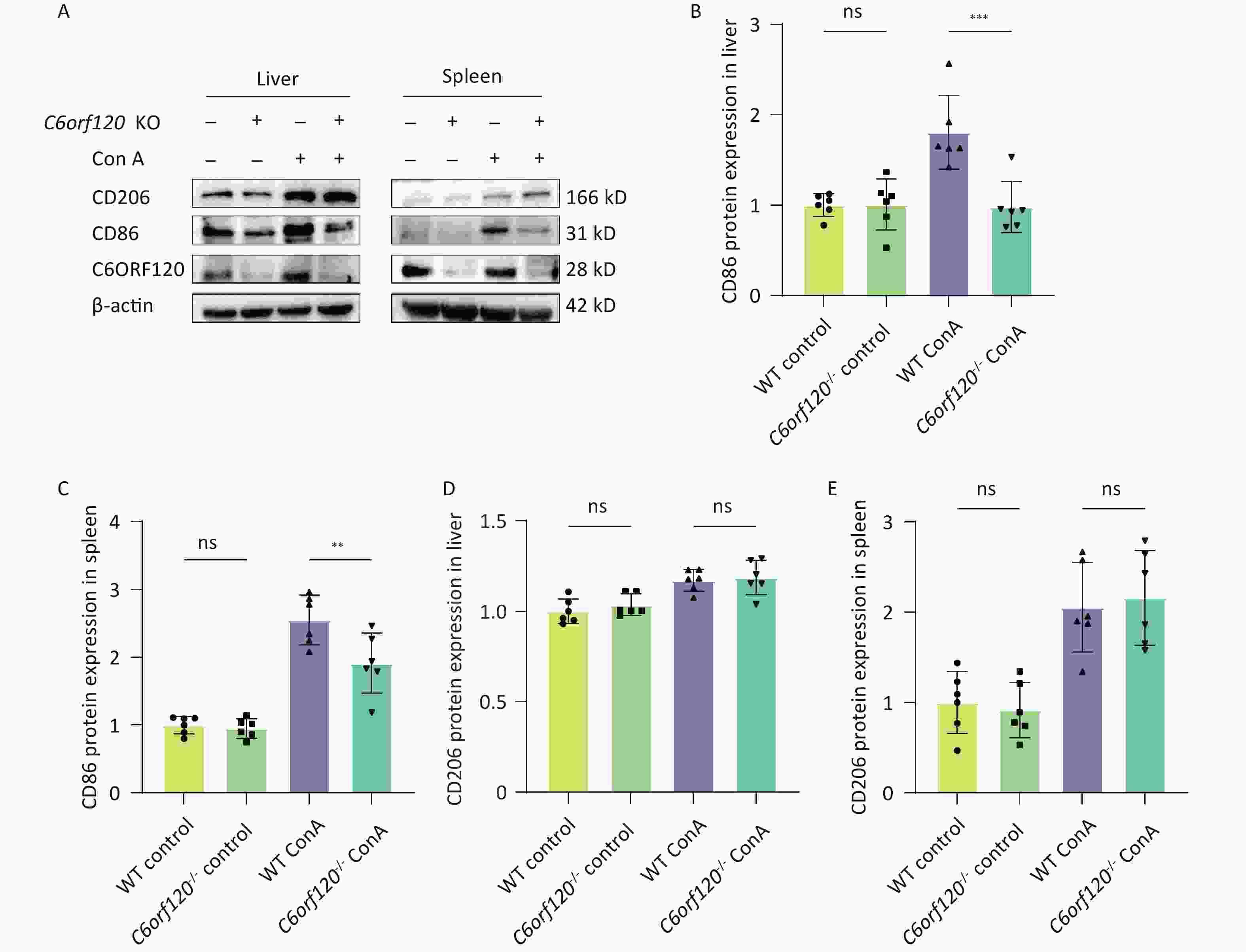
Figure 4. Protein expression of CD86 and CD206. (A) Western blotting analysis of liver and spleen tissues. Pixel intensity plots; CD86 protein expression in the liver (B) and spleen (C) (n = 6); CD206 protein expression in the liver (D) and spleen (E) (n = 6). **P < 0.01; *** P < 0.001; WT, wild-type; ns, no statistical significance.
-
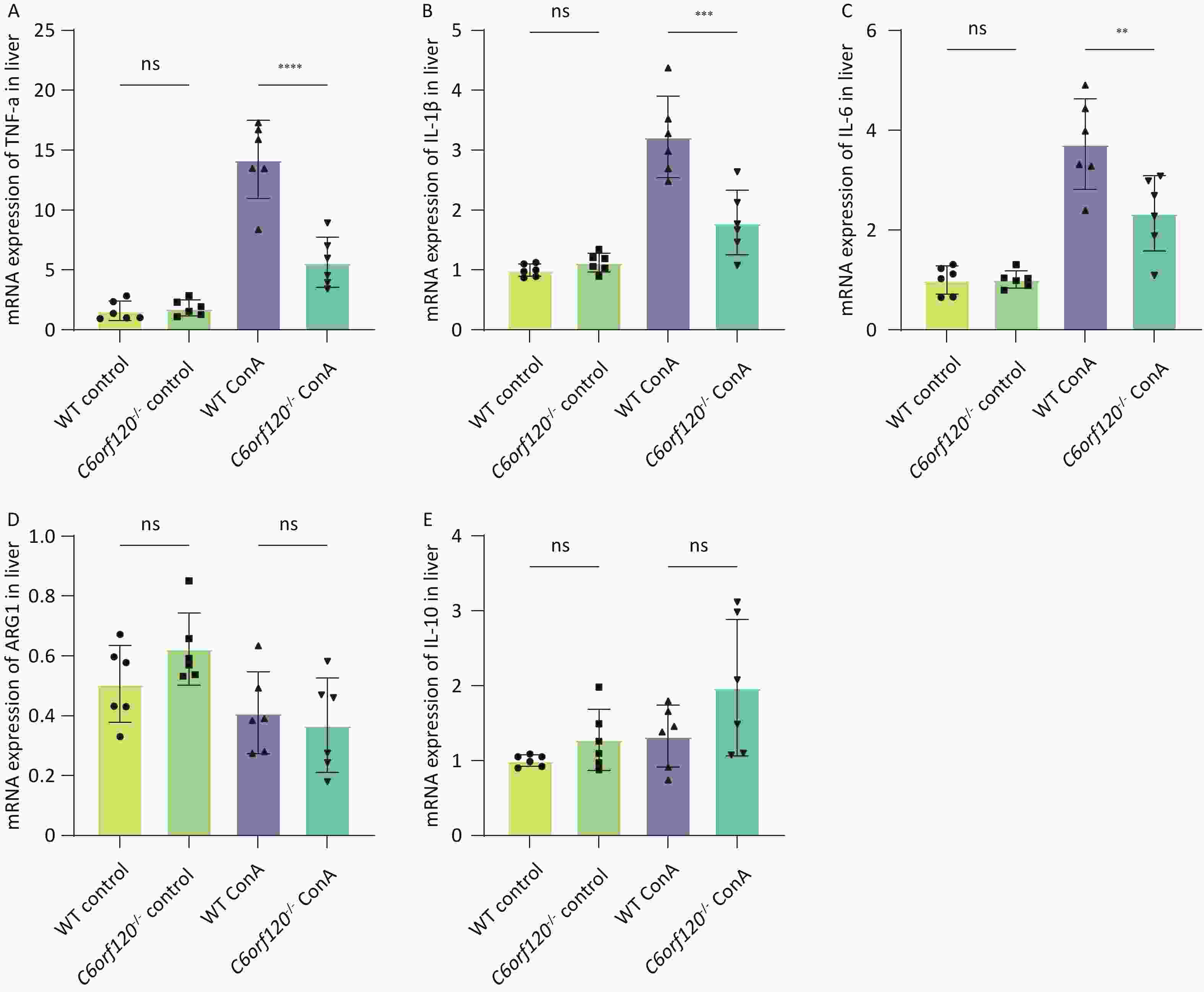
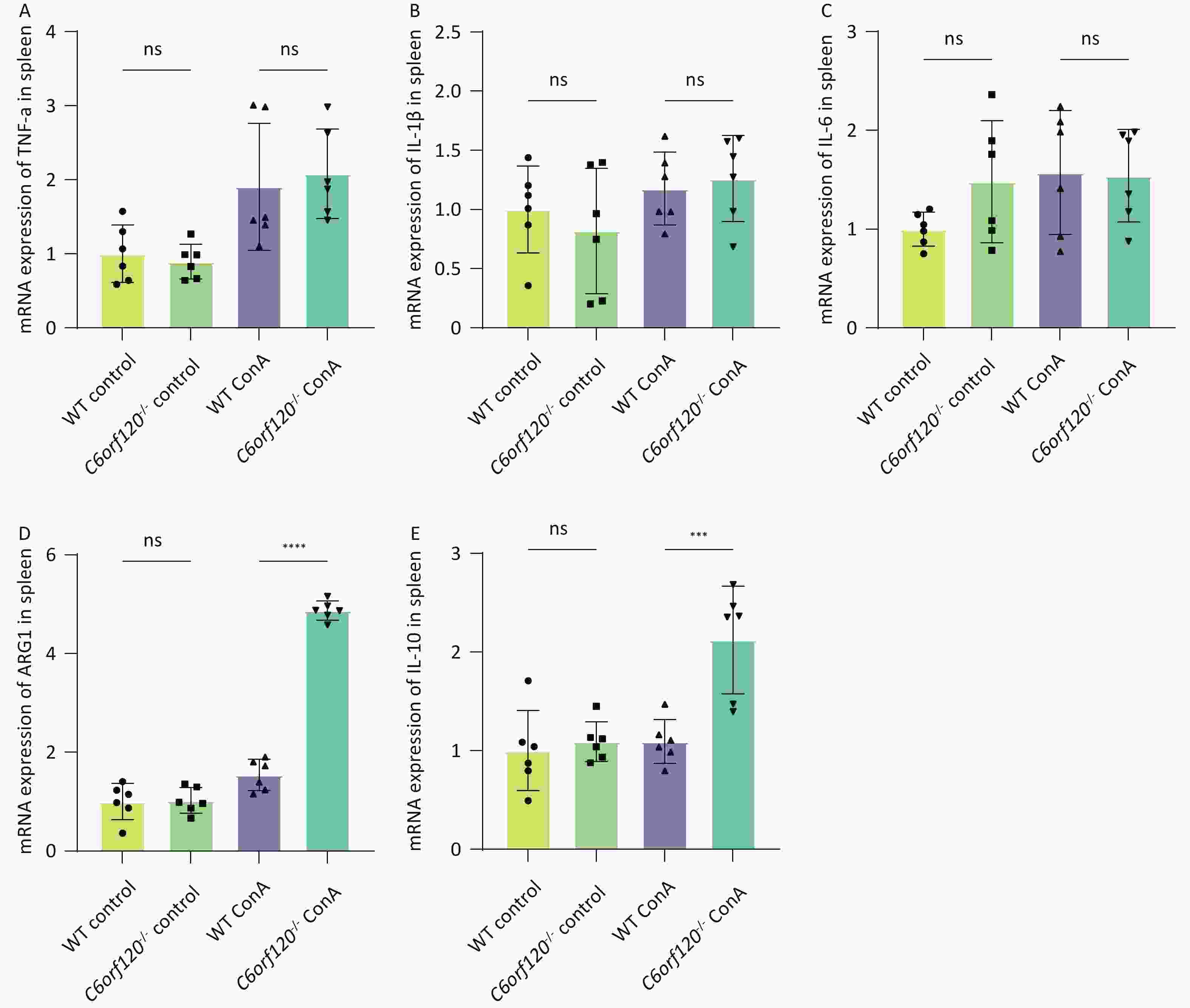
To investigate in depth the effect of C6orf120 deletion on the levels of inflammatory factors, we extracted total mRNA from liver and spleen tissues and used RT-qPCR to estimate the expression of TNF-α, IL-1β, and IL-6 pro-inflammatory factors, which are also markers of macrophage polarization toward the M1 phenotype. Additionally, the expression of ARG1 and IL-10 were detected, as markers of macrophage polarization toward the M2 type. The statistical analyses confirmed that the absence of C6orf120 reduced the levels of the pro-inflammatory factors TNF-α, IL-1β, and IL-6 in liver tissues in the AIH model, which is consistent with our previous findings (Figure 5A–E). However, in the spleen, we did not observe upregulation of the expression of M1-associated inflammatory factors, but upregulation of the expression of the M2-associated molecules ARG1 and IL-10 (Supplementary Figure S1A–E, available in www.besjournal.com).
-
THP-1 cells, a human macrophage cell line, were used for in vitro experiments[18]. First, the suspended macrophages were stimulated with PMA to become polarization-competent initial M0-adherent cells, which characteristically express the macrophage marker CD68 (Figure 6A)[19]. M0 macrophages were polarized to M1 macrophages (CD68+CD80+) by adding LPS and IFN-γ for 48 h. With IL4 and IL13 for 48–72 h, M0 macrophages were polarized into M2 macrophages (CD68+CD206+) (Figure 6B)[20]. Subsequently, to investigate in depth the effect of the C6orf120 gene on macrophage polarization, C6orf120-knockdown THP-1 cells were successfully generated (Figure 6C). C6orf120 knockdown did not affect the proportion of M1 and M2 macrophages and the expression of associated RNAs, including CD80, TNF-α, and CD206 (Figure 6D–G). Altogether, the above results indicate that gene knockdown in THP-1 cells does not affect macrophage polarization.
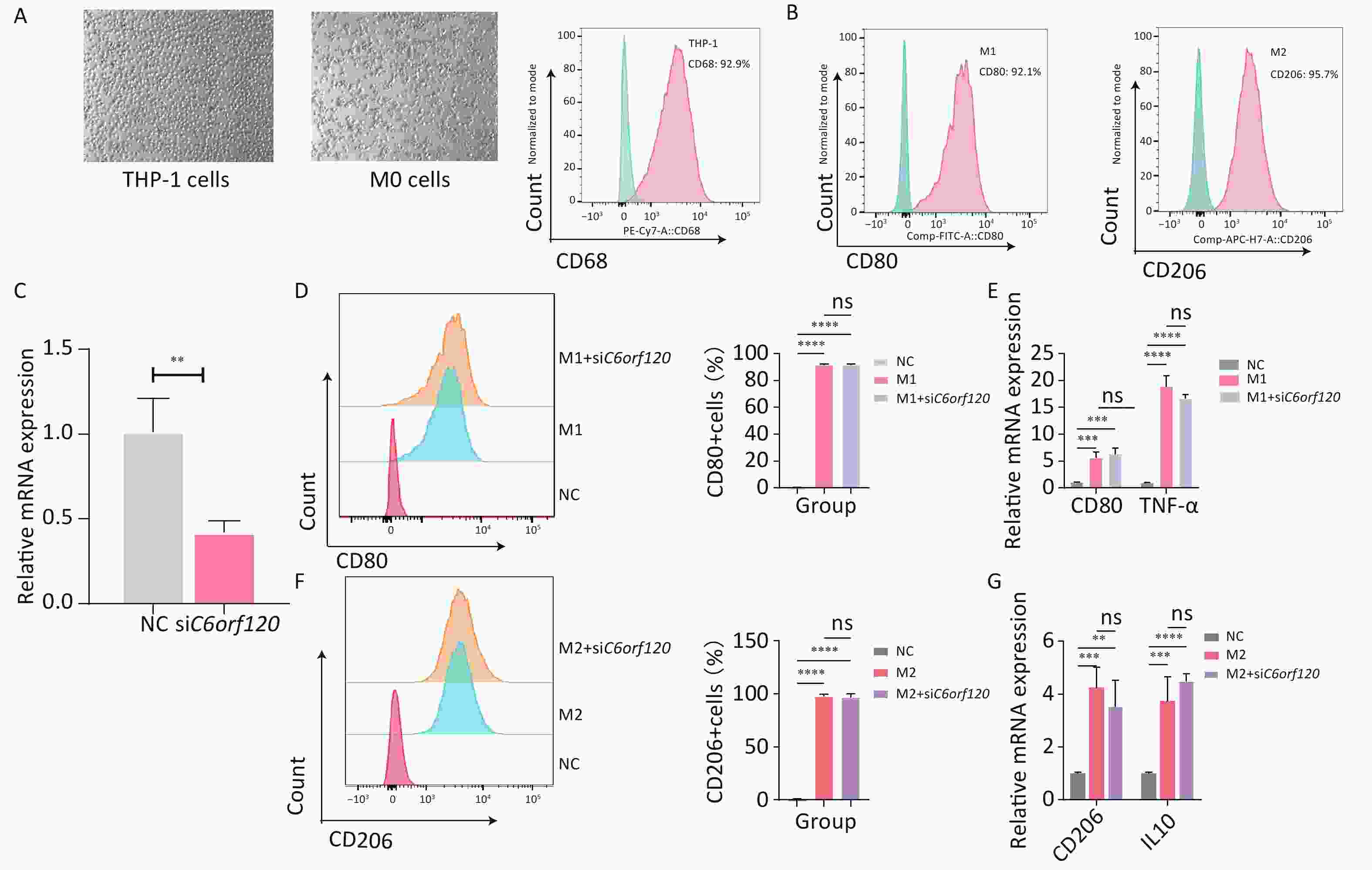
Figure 6. C6orf120 knockdown did not affect THP-1 cell polarization. (A) Pictures of PMA-induced originally suspended THP-1 cells becoming wall-adherent M0 cells and flow cytometry analysis of the proportion of CD68+ M0 cells. (B) Flow cytometry analysis of CD68+CD80+M1 macrophage and CD68+CD206+M2 macrophage proportions. (C) RT-qPCR analysis of C6orf120 mRNA expression. (D) Flow cytometry analysis of the proportion of M1 macrophages (CD68+CD80+) induced by IFN-γ and LPS in C6orf120 knockout versus non-knockout THP-1 cells. (E) RT-qPCR analysis of CD80 and TNF-α mRNA levels. (F) Flow cytometry analysis of the proportion of M2 macrophages (CD68+CD206+) induced by IL4 and IL13 in C6orf120 knockout versus non-knockout THP-1 cells. (G) RT-qPCR analysis of CD206 and IL10 mRNA levels. **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns represents no statistical significance.
-
Previous studies have suggested that the end product of the C6orf120 gene is a secreted glycosylated protein and that it may function in this form[12]. Therefore, we synthesized the recombinant C6ORF120 protein (rC6ORF120). Different concentrations of rC6ORF120 were added to the M0 cell culture medium to observe its effect on M0 cell polarization. We found that M0 cells would be polarized toward the M1 pro-inflammatory direction and the expression of associated RNAs CD80, TNF-α, and IL6 increased significantly upon stimulation with rC6ORF120 at a concentration of 500 ng/mL (Figure 7A–C). To verify whether C6ORF120 has a synergistic or antagonistic effect, we added 500 ng/mL of rC6ORF120 to THP-1 cells that had been successfully polarized into M1 and M2 macrophages. We found that the recombinant protein did not exacerbate the pro-inflammatory effects of LPS and IFN-γ, but reduced the frequency of IL4- and IL13-induced M2 macrophages as well as CD206 mRNA expression (Figure 7D–G). Taken together, the experimental results demonstrated the pro-inflammatory effect of rC6ORF120 in promoting macrophage polarization toward M1 macrophages.
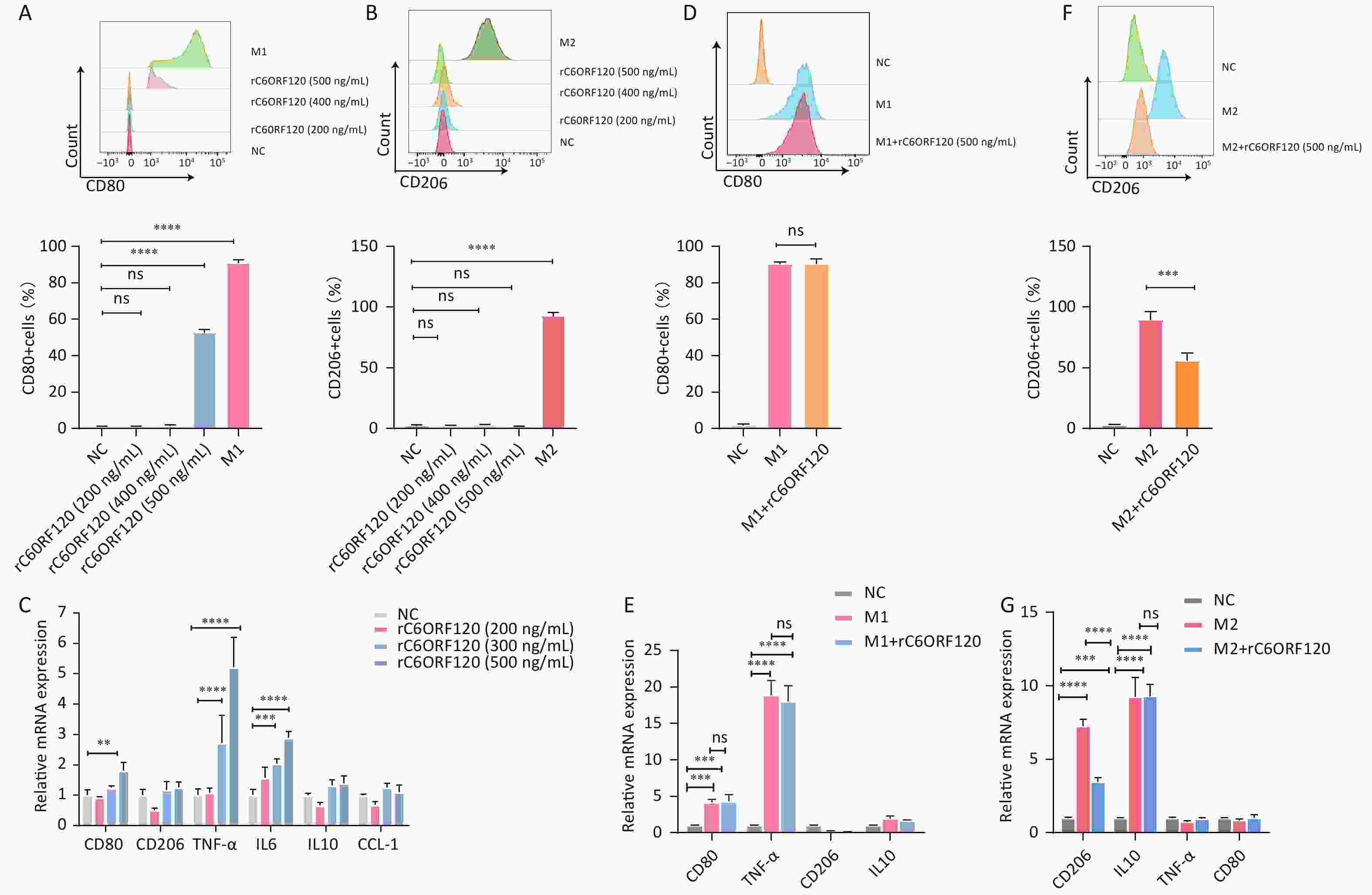
Figure 7. rC6ORF120 exerted pro-inflammatory effects on THP-1 cells. The proportions of CD68+CD80+M1 macrophages (A) and CD68+CD206+M2 macrophages (B) were determined using flow cytometry in the presence of different concentrations of rC6ORF120. (C) RT-qPCR analysis of mRNA levels of CD80, CD206, TNF-α, IL-6, IL-10, and CCL-1. (D) Flow cytometry analysis of the effect of rC6ORF120 on the proportion of LPS + IFN-γ-induced M1 macrophages and (E) RT-qPCR analysis of CD80, CD206, TNF-α, and IL-10 mRNA levels. (F) Flow cytometry analysis of the effect of rC6ORF120 on the proportion of IL4+IL13-induced M2 macrophages and (E) RT-qPCR analysis of CD80, CD206, TNF-α, and IL-10 mRNA levels. **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns represents no statistical significance.
-
AIH is an inflammatory disease in which abnormal immune function is the main etiological mechanism[21]. In addition to T-lymphocyte disorders, the pro-inflammatory phenotype of macrophages — M1 macrophages — is strongly associated with immune damage in AIH[6,22]. Previous studies have demonstrated that C6orf120 elimination reduces autoimmune liver injury by regulating the function of a variety of immune cells, including T and NKT cells[13-14]. However, potential influences on macrophages have not been explored.
In this study, we discovered that C6ORF120 protein expression was higher in the livers of rats and the serum of patients with AIH, suggesting that C6orf120 is closely associated with AIH (Figure 2A). The low levels of ALT and AST and the histological scores in the C6orf120-/- rats in this study corroborated the finding that C6orf120 deletion is beneficial for AIH (Figure 2). In addition, for the first time, C6orf120 knockout was found to inhibit the polarization of rat liver macrophages toward the pro-inflammatory phenotype and suppress the expression of related inflammatory factors, finally leading to attenuation of AIH (Figure 3). In in vitro experiments, rC6ORF120 similarly promoted the polarization of M0 cells to M1 macrophages (Figure 7). This finding connects C6orf120 to the polarization function of macrophages, providing a potential therapeutic target for the treatment of AIH.
Liver macrophages, the body's immune frontline cells against external pathogens, play a key role in maintaining hepatic homeostasis and tissue injury response and are involved in various liver diseases[23-25]. Diversity and plasticity characterize macrophages[26]. Macrophages can polarize into different cell subpopulations, predominantly M1 and M2 macrophages, and release associated cytokines[27]. Moreover, M1 and M2 macrophages can be dynamically transformed both in vivo and in vitro[28]. Pathological damage in AIH is often accompanied by infiltration of inflammatory macrophages[19]. In vivo, the experiments demonstrated that the absence of C6orf120 in the liver of AIH rats was accompanied by a decrease in the frequency of M1 macrophages and the secretion of the pro-inflammatory cytokines TNF-α, IL-1β, and IL-6, compared with WT rats (Figures 4–5). This confirmed the significant role of C6orf120 in the regulation of macrophage polarization.
Macrophage polarization can be affected by multiple factors, such as microorganisms, tissue microenvironment, and cytokines[24]. As important cell components in the complex immune microenvironment, cytokines strongly influence macrophage polarization[29]. M1 macrophages can be activated by Toll-like receptor ligands such as LPS and GM-CSF, or various Th1-type cytokines such as IFN-γ[30]. Zhang and Liu demonstrated that C6orf120 is involved in the regulation of T-cell apoptosis[14,31]. Wu et al. confirmed that C6orf120 knockdown increased Treg cell frequency and decreased the expression of IFN-γ from T and NKT cells[13]. Hence, we speculated that, in the organism, the effect of C6orf120 on the T-cell population and its cytokines may be one of the reasons for the effect of C6orf120 on the pro-inflammatory polarization of macrophages. Furthermore, in the THP-1 cell experiments, the presence of rC6ORF120 increased the frequency of M1 macrophages with the mRNA expression of the marker CD80 and decreased the frequency of M2 macrophages with the mRNA expression of the marker CD206. At the same time, the levels of the pro-inflammatory factors TNF-α and IL-6 were elevated (Figure 7). All these findings predict that the C6ORF120 protein may be a novel pro-inflammatory factor or molecule. However, the effects of C6orf120 on the immune microenvironment may be multifaceted, and its role in immunomodulation still needs to be thoroughly investigated.
The exact mechanism of C6orf120 regulation of macrophage polarization is unclear. The NF-κB/PI-3 and JAK/STAT pathways are classic macrophage polarization-related pathways[32]. The STAT family comprises various isoforms, of which STAT1, STAT3, and STAT6 are considered to be associated with liver inflammation. JAK binding by the cytokine receptor leads to receptor activation and phosphorylation of tyrosine residues in the receptor tail by JAK, which subsequently phosphorylates STAT downstream, ultimately inducing gene transcription[33]. Activation of the macrophage JAK1/STAT1 pathway is markedly a pro-inflammatory event, with a significant increase in the number of M1 macrophages, whereas activation of STAT6 inhibits NF-κB-mediated pro-inflammatory effects[34]. STAT3 is thought to contribute to the polarization of M2 macrophages in response to p-JAK3 activation and play a protective role in liver inflammation models[35]. However, some studies have suggested a pro-inflammatory role for STAT3 in the liver through crosstalk between various factors[36]. In a previous study, C6orf120-/- rats exhibited lower expression of p-JAK1, p-JAK3, its downstream p-STAT1, and p-STAT3 in the liver[13]. However, there was no significant difference in NF-κB expression in western blots[37]. The role of C6orf120 in the regulation of M1-cell polarization may be related to JAK/STAT.
In recent years, a variety of drugs and compounds have been identified that ameliorate liver disease by modulating macrophage polarization; many of these act by regulating the expression of non-coding RNAs[38-40]. Thus, manipulating the expression of lncRNAs and miRNAs may be a novel therapeutic modality. MicroRNA17-5p was found to attenuate collagenous arthritis by targeting the JAK-STAT pathway to reduce the number of inflammatory macrophages[41]. Meanwhile, Lynam-Lennon et al. concluded that C6orf120 is one of the target genes of microRNA17-5p and that they are negatively correlated[42]. We hypothesize that microRNA17-5p is likely to be a potential drug target for the regulation of C6orf120. In addition, as experiments have shown that rC6ORF120 can exert a pro-inflammatory effect, drugs or compounds that would neutralize C6ORF120 proteins may hold the key to future therapies, providing a wide range of ideas for the treatment of AIH.
This study has some limitations. First, the mechanisms regulating macrophage polarization are complex, and the mechanism of macrophage polarization by C6orf120 needs to be explored in depth. Next, the possibility that C6orf120 affects AIH by regulating other immune cells and cytokines requires further experimental confirmation. In addition, the association of C6orf120 with clinical patients with AIH and the development of clinical drugs should be the future focus of research.
In conclusion, C6orf120 knockout alleviated Con A-induced AIH in rats by inhibiting macrophage polarization toward M1 macrophages and by decreasing the level of pro-inflammatory factors TNF-α, IL-1β, and IL-6. In addition, rC6ORF120 protein stimulated macrophage polarization in a pro-inflammatory direction. Based on this study, C6orf120 shows promise as a target for AIH treatment in clinical practice.
-
Xin Wang wrote the article; Xin Wang, Yuqi Wang, and Hui Liu performed the experiments; Yingying Lin, Peng Wang, and Yunyun Yi assisted in animal experiments and primary cell extraction; Yuqi Wang assisted in data analysis; and the corresponding author Xin Li reviewed and corrected the article.
doi: 10.3967/bes2024.066
Knockout of C6orf120 in Rats Alleviates Concanavalin A-induced Autoimmune Hepatitis by Regulating Macrophage Polarization
-
Abstract:
Objective The effect of the functionally unknown gene C6orf120 on autoimmune hepatitis was investigated on C6orf120 knockout rats (C6orf120-/-) and THP-1 cells. Method Six–eight-week-old C6orf120-/- and wild-type (WT) SD rats were injected with Con A (16 mg/kg), and euthanized after 24 h. The sera, livers, and spleens were collected. THP-1 cells and the recombinant protein (rC6ORF120) were used to explore the mechanism in vitro. The frequency of M1 and M2 macrophages was analyzed using flow cytometry. Western blotting and PCR were used to detect macrophage polarization-associated factors. Results C6orf120 knockout attenuated Con A-induced autoimmune hepatitis. Flow cytometry indicated that the proportion of CD68+CD86+M1 macrophages from the liver and spleen in the C6orf120-/- rats decreased. C6orf120 knockout induced downregulation of CD86 protein and the mRNA levels of related inflammatory factors TNF-α, IL-1β, and IL-6 in the liver. C6orf120 knockout did not affect the polarization of THP-1 cells. However, rC6ORF120 promoted the THP-1 cells toward CD68+CD80+M1 macrophages and inhibited the CD68+CD206+M2 phenotype. Conclusion C6orf120 knockout alleviates Con A-induced autoimmune hepatitis by inhibiting macrophage polarization toward M1 macrophages and reducing the expression of related inflammatory factors in C6orf120-/- rats. -
Key words:
- C6orf120 /
- Autoimmune hepatitis /
- Macrophage polarization /
- M1 macrophages
注释: -
Figure 2. Assessment of liver injury. (A) Western blotting analyzing C6ORF120 protein in the liver tissue in the presence or absence of Con A stimulation; ELISA analyzing C6ORF120 protein level in healthy volunteers and patients with AIH. (B) H & E staining of liver sections (black arrows indicate inflammatory cell infiltration; green arrows, fatty degeneration; and blue arrows, necrosis). (C) Histopathological score of the liver. (D, E) Serum ALT and AST levels (n = 6). *P < 0.05; ***P < 0.001; and ****P < 0.0001. WT, wild-type; ns, no statistical significance.
Figure 3. Macrophage subsets in vivo. (A) Representative image of the gating strategy of CD68+CD86+M1 macrophages in liver and spleen lymphocytes. Frequency of M1 macrophages in the liver (B) and spleen (C). (D) Representative images of the gating strategy of CD68+CD163+M2 macrophages in liver and spleen lymphocytes. Frequency of M2 macrophages in the liver (E) and spleen (F) (n = 6). **P < 0.01; ****P < 0.0001; WT, wild-type; ns, no statistical significance.
Figure 4. Protein expression of CD86 and CD206. (A) Western blotting analysis of liver and spleen tissues. Pixel intensity plots; CD86 protein expression in the liver (B) and spleen (C) (n = 6); CD206 protein expression in the liver (D) and spleen (E) (n = 6). **P < 0.01; *** P < 0.001; WT, wild-type; ns, no statistical significance.
Figure 6. C6orf120 knockdown did not affect THP-1 cell polarization. (A) Pictures of PMA-induced originally suspended THP-1 cells becoming wall-adherent M0 cells and flow cytometry analysis of the proportion of CD68+ M0 cells. (B) Flow cytometry analysis of CD68+CD80+M1 macrophage and CD68+CD206+M2 macrophage proportions. (C) RT-qPCR analysis of C6orf120 mRNA expression. (D) Flow cytometry analysis of the proportion of M1 macrophages (CD68+CD80+) induced by IFN-γ and LPS in C6orf120 knockout versus non-knockout THP-1 cells. (E) RT-qPCR analysis of CD80 and TNF-α mRNA levels. (F) Flow cytometry analysis of the proportion of M2 macrophages (CD68+CD206+) induced by IL4 and IL13 in C6orf120 knockout versus non-knockout THP-1 cells. (G) RT-qPCR analysis of CD206 and IL10 mRNA levels. **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns represents no statistical significance.
Figure 7. rC6ORF120 exerted pro-inflammatory effects on THP-1 cells. The proportions of CD68+CD80+M1 macrophages (A) and CD68+CD206+M2 macrophages (B) were determined using flow cytometry in the presence of different concentrations of rC6ORF120. (C) RT-qPCR analysis of mRNA levels of CD80, CD206, TNF-α, IL-6, IL-10, and CCL-1. (D) Flow cytometry analysis of the effect of rC6ORF120 on the proportion of LPS + IFN-γ-induced M1 macrophages and (E) RT-qPCR analysis of CD80, CD206, TNF-α, and IL-10 mRNA levels. (F) Flow cytometry analysis of the effect of rC6ORF120 on the proportion of IL4+IL13-induced M2 macrophages and (E) RT-qPCR analysis of CD80, CD206, TNF-α, and IL-10 mRNA levels. **P < 0.01; ***P < 0.001; ****P < 0.0001; and ns represents no statistical significance.
Table 1. Primer sequence for RT-qPCR
Gene (ID) Species Forward (5‘-3’) Reverse (5‘-3’) GAPDH Rat GGCATCGTGGAAGGGCTCAT CGTCGGGTCTTGTAGTAGGGA TNF-α Rat GCGATGTGGAACTGGCAGAGG GAGAAGAGTAAGGACGAGCACCG IL-1β Rat ATCTCACAGCAGCATCTCGACAAG CCTACTACTGCTGGACGATCACAC IL-6 Rat TTCCAGCCAGTTGCCTTCTT GTGAAGTGTTCAGCCTCCGAA ARG1 Rat CCAAGCCAAAGCCCATAGAGAT ACCAGGCCAGCTTTCCTTAAT IL-10 Rat GAAGGACCAGCTGGACAACA GGGGCATCACTTCTACCAGG GAPDH Human AGAAGGCTGGGGCTCATTTG AGGGGCCATCCACAGTCTTC CCL1 Human CTCATTTGCGGAGCAAGAGAT GCCTCTGAACCCATCCAACTG IL6 Human TGGCAGAAAACAACCTGAACC GGCTTGTTCCTCACTACTCTCA IL10 Human GGCATCTACAAAGCCATGAGTG TTTCTCAAGGGGCTGGGTCA CD206 Human GGGACGTGGCTGTGGATAAA TCCAAAACCCAGAAGACGCA TNF-α Human TGCACTTTGGAGTGATCGGC ACTCGGGGTTCGAGAAGATG CD80 Human TTGGTGCTGGCTGGTCTTTC TGCCAGTAGATGCGAGTTTGT -
[1] Obermayer-Straub P, Strassburg CP, Manns MP. Autoimmune hepatitis. J Hepatol, 2000; 32, 181−97. [2] Mieli-Vergani G, Vergani D, Czaja AJ, et al. Autoimmune hepatitis. Nat Rev Dis Primers, 2018; 4, 18017. doi: 10.1038/nrdp.2018.17 [3] Lamba M, Ngu JH, Stedman CAM. Trends in incidence of autoimmune liver diseases and increasing incidence of autoimmune hepatitis. Clin Gastroenterol Hepatol, 2021; 19, 573-79. e1. [4] Heymann F, Hamesch K, Weiskirchen R, et al. The concanavalin A model of acute hepatitis in mice. Lab Anim, 2015; 49, 12−20. doi: 10.1177/0023677215572841 [5] Hao JH, Sun WL, Xu HC. Pathogenesis of Concanavalin A induced autoimmune hepatitis in mice. Int Immunopharmacol, 2022; 102, 108411. doi: 10.1016/j.intimp.2021.108411 [6] Chi G, Pei JH, Li XQ. EZH2-mediated H3K27me3 promotes autoimmune hepatitis progression by regulating macrophage polarization. Int Immunopharmacol, 2022; 106, 108612. doi: 10.1016/j.intimp.2022.108612 [7] Liu Y, Liu H, Zhu JS, et al. Interleukin-34 drives macrophage polarization to the M2 phenotype in autoimmune hepatitis. Pathol -Res Pract, 2019; 215, 152493. doi: 10.1016/j.prp.2019.152493 [8] Grønbæk H, Kreutzfeldt M, Kazankov K, et al. Single-centre experience of the macrophage activation marker soluble (s)CD163 - associations with disease activity and treatment response in patients with autoimmune hepatitis. Aliment Pharmacol Ther, 2016; 44, 1062−70. doi: 10.1111/apt.13801 [9] Roohani S, Tacke F. Liver injury and the macrophage issue: molecular and mechanistic facts and their clinical relevance. Int J Mol Sci, 2021; 22, 7249. doi: 10.3390/ijms22147249 [10] Guilliams M, Scott CL. Liver macrophages in health and disease. Immunity, 2022; 55, 1515−29. doi: 10.1016/j.immuni.2022.08.002 [11] Guo LP, Zhou L, Li HX, et al. The study of liver macrophages polarization in patients with autoimmune hepatitis. Chin J Intern Med, 2017; 56, 763−5. (In Chinese [12] Li X, Qiao Y, Chang LS, et al. Role of C6ORF120, an N-glycosylated protein, is implicated in apoptosis of CD4+ T lymphocytes. Chin Med J, 2011; 124, 3560−7. [13] Wu YN, Zhang R, Song XC, et al. C6orf120 gene knockout in rats mitigates concanavalin A-induced autoimmune hepatitis via regulating NKT cells. Cell Immunol, 2022; 371, 104467. doi: 10.1016/j.cellimm.2021.104467 [14] Zhang MK, Ma HM, Zhang J, et al. Deletion of the C6orf120 gene with unknown function ameliorates autoimmune hepatitis induced by concanavalin A. Cell Immunol, 2018; 331, 9−15. doi: 10.1016/j.cellimm.2018.04.017 [15] National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the care and use of laboratory animals. 8th ed. National Academies Press (US). 2011. [16] Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol, 1995; 22, 696−9. doi: 10.1016/0168-8278(95)80226-6 [17] Orme J, Mohan C. Macrophage subpopulations in systemic lupus erythematosus. Discov Med, 2012; 13, 151−8. [18] Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol, 2014; 23, 37−45. doi: 10.1016/j.intimp.2014.08.002 [19] Wang C, Ma C, Gong LH, et al. Macrophage polarization and its role in liver disease. Front Immunol, 2021; 12, 803037. doi: 10.3389/fimmu.2021.803037 [20] Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity, 2014; 41, 14−20. doi: 10.1016/j.immuni.2014.06.008 [21] Sucher E, Sucher R, Gradistanac T, et al. Autoimmune hepatitis-immunologically triggered liver pathogenesis-diagnostic and therapeutic strategies. J Immunol Res, 2019; 2019, 9437043. [22] Han RR, Xiao JT, Zhai H, et al. Dimethyl fumarate attenuates experimental autoimmune neuritis through the nuclear factor erythroid-derived 2-related factor 2/hemoxygenase-1 pathway by altering the balance of M1/M2 macrophages. J Neuroinflammation, 2016; 13, 97. doi: 10.1186/s12974-016-0559-x [23] Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci, 2021; 22, 6995. doi: 10.3390/ijms22136995 [24] Funes SC, Rios M, Escobar-Vera J, et al. Implications of macrophage polarization in autoimmunity. Immunology, 2018; 154, 186−95. doi: 10.1111/imm.12910 [25] Zhang J, Muri J, Fitzgerald G, et al. Endothelial lactate controls muscle regeneration from ischemia by inducing M2-like macrophage polarization. Cell Metab, 2020; 31, 1136-53. e7. [26] Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest, 2012; 122, 787−95. doi: 10.1172/JCI59643 [27] Orecchioni M, Ghosheh Y, Pramod AB, et al. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively activated macrophages. Front Immunol, 2019; 10, 1084. doi: 10.3389/fimmu.2019.01084 [28] Liu YC, Zou XB, Chai YF, et al. Macrophage polarization in inflammatory diseases. Int J Biol Sci, 2014; 10, 520−9. doi: 10.7150/ijbs.8879 [29] Muñoz J, Akhavan NS, Mullins AP, et al. Macrophage polarization and osteoporosis: a review. Nutrients, 2020; 12, 2999. doi: 10.3390/nu12102999 [30] Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity, 2010; 32, 593−604. doi: 10.1016/j.immuni.2010.05.007 [31] LIU H, WANG X, WANG P, et al. Novel protein C6ORF120 promotes apoptosis through mitochondria-dependent pathway in CD4+T lymphocytes. Biomed Environ Sci, 2023; 36, 639−43. [32] Kerneur C, Cano CE, Olive D. Major pathways involved in macrophage polarization in cancer. Front Immunol, 2022; 13, 1026954. doi: 10.3389/fimmu.2022.1026954 [33] Xin P, Xu XY, Deng CJ, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol, 2020; 80, 106210. doi: 10.1016/j.intimp.2020.106210 [34] Huang IH, Chung WH, Wu PC, et al. JAK-STAT signaling pathway in the pathogenesis of atopic dermatitis: an updated review. Front Immunol, 2022; 13, 1068260. doi: 10.3389/fimmu.2022.1068260 [35] Bode JG, Ehlting C, Häussinger D. The macrophage response towards LPS and its control through the p38MAPK-STAT3 axis. Cell Signal, 2012; 24, 1185−94. doi: 10.1016/j.cellsig.2012.01.018 [36] Qin HW, Holdbrooks AT, Liu YD, et al. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J Immunol, 2012; 189, 3439−48. doi: 10.4049/jimmunol.1201168 [37] Zhang J, Zhang MK, Ma HM, et al. C6orf120 gene deficiency may be vulnerable to carbon tetrachloride induced acute hepatic injury in rats. Arch Med Sci, 2020; 18, 1626−37. [38] Hu F, Tong JK, Deng BL, et al. MiR-495 regulates macrophage M1/M2 polarization and insulin resistance in high-fat diet-fed mice via targeting FTO. Pflugers Arch- Eur J Physiol, 2019; 471, 1529−37. doi: 10.1007/s00424-019-02316-w [39] Xiao T, Zou ZL, Xue JC, et al. LncRNA H19-mediated M2 polarization of macrophages promotes myofibroblast differentiation in pulmonary fibrosis induced by arsenic exposure. Environ Pollut, 2021; 268, 115810. doi: 10.1016/j.envpol.2020.115810 [40] Wang YH, Smith W, Hao DJ, et al. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int Immunopharmacol, 2019; 70, 459−66. doi: 10.1016/j.intimp.2019.02.050 [41] Najm A, Masson FM, Preuss P, et al. MicroRNA-17-5p reduces inflammation and bone erosions in mice with collagen-induced arthritis and directly targets the JAK/STAT pathway in rheumatoid arthritis fibroblast-like synoviocytes. Arthritis Rheumatol, 2020; 72, 2030−9. doi: 10.1002/art.41441 [42] Lynam-Lennon N, Heavey S, Sommerville G, et al. MicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotype. Oncotarget, 2017; 8, 11400−13. doi: 10.18632/oncotarget.13940 -




 下载:
下载:



 Quick Links
Quick Links