-
Infections by Cronobacter spp. can result in a range of symptoms among different age groups. Compared with the adult population, a higher percentage of invasive infections occur among children less than five years of age[9]. These infections are hazardous to infants since they can lead to neonatal meningitis, bacteremia, and necrotizing enterocolitis (NEC). Based on multi-locus sequence typing (MLST), the genus Cronobacter consists of seven species, replacing the former single species Enterobacter sakazakii classification. There are many sequence types (STs) of Cronobacter. Many cases of neonatal meningitis are caused by Cronobacter spp. ST 4 of C. sakazakii[3]. The source of Cronobacter infections remains unclear since strains have been isolated from throat and sputum samples as well as from rectal and fecal swabs[1]. Although Cronobacter spp. exist widely in nature, including food, plants, water, and soil, as well as in housekeeping environments, they are recognized as a public health threat because they cause disease in cases of powdered infant formula (PIF) contamination.
Bacterial antibiotic resistance is a serious problem in treating hospital infections, thus surveillance of antibiotic susceptibility profiles is necessary. At the end of the 20th century, Cronobacter spp. were reported to be more sensitive to some antibiotics than other Enterobacter species[2]. Unfortunately, it was soon determined that Cronobacter spp. were resistant to broad-spectrum penicillins and to cephalosporins through the production of β-lactamases[5]. Recently, like other Enterobacteriaceae, Cronobacter spp. were frequently found to be resistant to β-lactam derivatives, macrolides, and aminoglycosides such as rifampicin, amoxicillin-clavulanic acid, streptomycin, tetracycline, or ampicillin. In addition, multi-resistant strains have also been detected[10]. The mechanism and source of their resistance to antibiotics are currently not well understood and are the focus of future studies.
In China, the isolation rate of Cronobacter spp. from commercial PIF or follow-up formula (FUF) samples is relatively high[8]. Nevertheless, clinical cases of Cronobacter infection have been ignored to date. This is the first report of two cases of Cronobacter infection of infants, identified at the Wuhan Women and Children Medical Care Center Hospital (Wuhan City, China). Here, we describe the cases, the genomic analysis of the isolates, and the antibiotic-resistance profiles of the two strains.
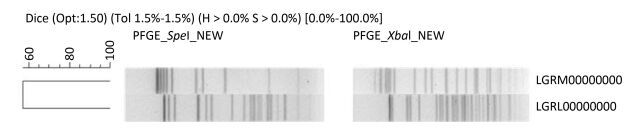
Case 1 Case 1 was a female infant born after 36 weeks of gestation (birth weight: 2900 g). Initially, the child appeared to be healthy and fed well on breast milk and supplementary PIF. 11 days after birth, she developed fever, muscle spasms in her extremities, and fed poorly. When admitted to hospital, her intracranial pressure was high and her spinal fluid contained numerous leukocytes. Treatment with intravenous cefoperazone, sulbactam sodium, meropenem, mannitol, and phenobarbital was started immediately. After a week, the infant was discharged from hospital and its mental and physical development was considered to be markedly impaired. Cronobacter was isolated from the cerebral spinal fluid (CSF) using selective medium (DFI agar). An API 20E kit was used for the presumptive identification of Cronobacter. Real-time PCR with primers for the macromolecular synthesis operon and MLST were performed for confirmation. By querying the MLST sequence type database for Cronobacter spp. (http://www.pubmlst.org/cronobacter/), the strain was identified as C. malonaticus ST 60. The strain was named Chcon_9 and was deposited in the Cronobacter MLST database. The pulse field gel electrophoresis (PFGE) pattern was compared with other patterns of the PulseNet China database, which contains strains (n = 252) mainly isolated from food, drinking water, and anal swabs. No matching pattern was found (Figure 1). This strain was tested for antibiotic susceptibility to 16 antibiotics using the broth dilution method and was found to be resistant to nine antibiotics including: ampicillin, azithromycin, ceftriaxone, chloramphenicol, doxycycline, gentamycin, tetracycline, trimethoprim-sulfamethoxazole, and sulfonamides. The minimal inhibitory concentration (MIC) values are shown in Table 1 and the results were interpreted using the Enterobacteriaceae data from the Clinical and Laboratory Standards Institute (CLSI). Whole genome sequencing was performed using an Illumina HiSeq 2500 sequencer and the assembled genome was compared with the Antibiotic Resistance Genes Database (ARDB; http://ardb.cbcb.umd.edu/index.html). Various genes known to encode antimicrobial resistance were identified within the genome sequence (Table 2). These whole genome results were deposited in DDBJ/EMBL/GenBank under the accession number LGRM00000000.

Figure 1. Dendrogram combining PFGE patterns of Xba I and Spe I digested DNA from Cronobacter spp. in this study.
Table 1. Minimum inhibitory concentrations of 16 antibiotics for Cronobacter in this Study
Antibiotics MIC (µg/mL) C. malonaticus HB03 C. sakazakii HB04 Ampicillin (AMP) ≥ 32 ≥ 3 Azithromycin (AZI) ≥ 64 16 Amoxicillin/clavulanic acid 2:1 ration (AUG2) 4/2 4/2 Cefoxitin (FOX) 2 ≥ 32 Cefepime (FEP) 4 ≥ 32 Ceftriaxone (AXO) ≥ 8 ≥ 8 Chloramphenicol (CHL) ≥ 64 ≥ 32 Ciprofloxacin (CIP) - 0.06 Doxycycline (DOX) ≥ 32 4 Gentamycin (GEN) ≥ 32 - Imipenem (IMI) - - Nalidixic acid (NAL) 2 4 Streptomycin (STR) 32 - Sulfonamides (SMX) ≥ 512 ≥ 512 Tetracycline (TET) ≥ 32 4 Trimethoprim-sulfamethoxazde (SXT) ≥ 4/75 0.25/4.75 Table 2. Putative antibiotic resistance genes and annotation of Cronobacter in this Study
Antibiotic Resistance Putative Gene Annotation Species C. malonaticus HB03 C. sakazaakii HB04 β-Lactam pbp1a
pbp1b
pbp2The enzyme has a penicillin-insensitive tranlycosylase N-terminal domain and a penicillin-insensitive transpeptidase C-teminal domain. +
+
++
+
+ampC
ampA, ampR
bl3_l
bl2be_
CTX-M-14
bl2be_shv2Class A, B, C β-lactamase. This enzyme breaks theβ-lactam antibiotic ring open and deactivates the molecule's antibacterial properties. +
+
+
-
++
-
+
+
-Aminoglycosides aac3iia Aminoglycoside N-acetyltransferase, which modifies aminoglycosides by acetylation. + - Macrolides aph3ia
aph33ib
aph6idAminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. +
+
+-
-
-Tetracycline ant2ia Aminoglycoside O-nucleotidylyltransferase, which modifies aminoglycosides by adenylylation. + - Phenicols erea Erythromycin esterase, which can inactivate erythromycin by lactone ring cleavage. + - tetd Major facilitator superfamily transporter, tetracycline efflux pump. + - tet34 Xanthine-guanine phosphoribosytransferase. + + cata2 Group A chloramphenicol acetyltransferase, which can inactivate chloramphenicol. + - Multidrug resistance efflux pump acra
acrb
emrd
emre
macb
mdtk
mdth
mdtg
oprm
tolc
ykkcMultidrug resistance efflux pump +
+
+
+
+
+
+
+
-
+
++
+
+
+
-
+
+
+
+
+
+Case 2 Case 2 was a female infant with no birth record details, as she was an abandoned baby living in a welfare house. She was fed with powdered-milk formula. When the baby was 10 days old, she had anoplasty because of an imperforate anus. After the operation, she started feeding poorly. Two months after birth, she developed anemia, diarrhea, moderate dehydration, and severe malnutrition. Although she was treated with anti-inflammatory drugs (intravenous meropenem), fluid replacement, and symptomatic treatments, her condition deteriorated. Finally, after 4 days in hospital, she died of circulatory and respiratory failure. Cronobacter was isolated from blood using DFI agar.
The strain was identified using the same methods as in Case 1. The PFGE pattern did not match the strain isolated in Case 1, or any entries in the PulseNet China database (Figure 1). The MLST profile of the strain was identified as C. sakazakii ST 83. The strain was named Chcon_10 and was deposited in the Cronobacter MLST database. The strain was resistant to the following six antibiotics: ampicillin, cefoxitin, ceftriaxone, cefepime, chloramphenicol, and sulfonamides (Table 1). Whole genome sequencing was performed as described in Case 1. The predicted antibiotic resistant genes are listed in Table 2. The whole genome results were deposited at DDBJ/EMBL/GenBank under the accession number LGRL00000000.
Seven species of Cronobacter have been identified, including C. sakazakii, C. malonaticus, C. dublinensis, C. turicensis, C. muytjensii, C. universalis, and C. condiment. However, C. sakazakii has mainly been reported to cause neonatal meningitis and NEC. In particular, C. sakazakii ST 4 has been more closely associated with neonatal meningitis[3]. In this study, the strain from Case 1 was C. malonaticus ST 60, which was firstly reported to cause neonatal meningitis. Although the other strain, identified as C. sakazakii ST 83, has been known to cause meningitis, it is uncommon in clinical practice. Contaminated PIF has been described as the primary cause of Cronobacter infection in feeding infants. However, we failed to trace the source of infection in these two cases, raising the possibility that there are other routes of transmission in Cronobacter infection.
Joshua B. Gurtler summarized the antibiotic resistance profile of Cronobacter infection and demonstrated that treatment should be guided by clinical judgment and in vitro antibiotic susceptibility testing[2]. Since then, although antibiotic resistance in clinical isolates has been reported less frequently, antibiotic resistance in isolates from food and the environment has been observed[7]. The most common antibiotics that Cronobacter spp. from food and the environment are resistant to were cephalothins and penicillins[10], in agreement with other reports[5]. The reason for this is that β-lactamase production appears to be more widespread among isolates of Cronobacter spp.[5]. Of the 16 antibiotics tested in this study, the two isolates were susceptible to amoxicillin/clavulanic acid, ciprofloxacin, imipenem, nalidixic acid, and streptomycin. Because antibiotic use is restricted in infants, carbapenems, such as imipenem, were actually the only choice for treatment. Multi-antibiotic resistance is common in Enterobacter spp., but rare in Cronobacter. Here, two cases with multi-antibiotic resistance are presented, indicating that there is a similar problem of antibiotic resistance for Cronobacter.
Whole genome sequencing identified the presence of various genes known to encode antibiotic resistance proteins. The presence of some of these genes, such as ampC, ampA, ampR, bl3_l, bl2be_CTX-M-14, and bl2be_shv2 is predicted to be related to β-lactamase-mediated antibiotic resistance. The unusual resistance phenotype in C. sakazakii and C. malonaticus has been attributed to AmpC β-lactamases[7]. Some Enterobacteriaceae, such as Serratia marcescens, Citrobacter freundii, Providencia spp. and Morganella morganii, that possess chromosomally determined AmpC β-lactamases, may express the enzymes at a high level following exposure to β-lactams. However, the risk of clinical failure using β-lactams that show susceptibility in vitro is less clear in these species than in Enterobacter spp.[4]. Although bl2be_CTX-M-14 and bl2be_shv2 have not been found in Cronobacter previously, the presence of CTX-M-15 and SHV-12 in Cronobacter has been reported[6]. In this study, many genes encoding multidrug resistance efflux pumps were screened. Bacterial multidrug resistance efflux pumps constitute an important mechanism of antibiotic resistance and are required by many pathogens for successful infection. For different antibiotics, different genes in the ARDB were found to be relevant (Table 2). Further studies are needed to determine whether the genes predicted in this study are functional.
This report highlights the importance of antibiotic resistance in two Cronobacter species that are known to cause life-threatening infections in infants. Treatment of Cronobacter infections with carbapenems or the newer third-generation cephalosporins in combination with an aminoglycoside or trimethoprim-sulfamethoxazole is recommended[6]. However, Cronobacter was not susceptible to third-generation cephalosporins, aminoglycoside, and trimethoprim-sulfamethoxazole in this study. As with Enterobacter spp., the antibiotic resistance of Cronobacter is a serious issue that needs more attention. Clinical studies that clearly define optimal treatment for Cronobacter spp. are required.
doi: 10.3967/bes2017.079
Two Cases of Multi-antibiotic Resistant Cronobacter spp.Infections of Infants in China
-
Abstract: Infections by Cronobacter spp. are hazardous to infants since they can lead to neonatal meningitis, bacteremia, and necrotizing enterocolitis. Cronobacter spp. are frequently resistant to β-lactam derivatives, macrolides, and aminoglycosides. In addition, multi-resistant strains have also been detected. In China, the isolation rate of Cronobacter spp. from commercial powdered infant formula (PIF) or follow-up formula (FUF) is relatively high. Nevertheless, clinical cases of Cronobacter infection have been ignored to date. Here we describe two cases of Cronobacter infection detected at the Wuhan Women and Children Medical Care Center Hospital (Wuhan City, China). We provide the genomic analysis of the isolates and the antibiotic-resistance profiles of the two strains. The Cronobacter strains identified in this study were not susceptible to third-generation cephalosporins, aminoglycoside, and/or trimethoprim-sulfamethoxazole. Whole genome sequencing revealed various genes known to encode antibiotic resistance. Future studies are needed to determine whether the genes predicted in this study are functional. As with Enterobacter spp., the antibiotic resistance of Cronobacter is a serious issue that requires more attention.
-
Key words:
- Multi-antibiotic resistant /
- Cronobacter spp. /
- Infant /
- Case report
-
Table 1. Minimum inhibitory concentrations of 16 antibiotics for Cronobacter in this Study
Antibiotics MIC (µg/mL) C. malonaticus HB03 C. sakazakii HB04 Ampicillin (AMP) ≥ 32 ≥ 3 Azithromycin (AZI) ≥ 64 16 Amoxicillin/clavulanic acid 2:1 ration (AUG2) 4/2 4/2 Cefoxitin (FOX) 2 ≥ 32 Cefepime (FEP) 4 ≥ 32 Ceftriaxone (AXO) ≥ 8 ≥ 8 Chloramphenicol (CHL) ≥ 64 ≥ 32 Ciprofloxacin (CIP) - 0.06 Doxycycline (DOX) ≥ 32 4 Gentamycin (GEN) ≥ 32 - Imipenem (IMI) - - Nalidixic acid (NAL) 2 4 Streptomycin (STR) 32 - Sulfonamides (SMX) ≥ 512 ≥ 512 Tetracycline (TET) ≥ 32 4 Trimethoprim-sulfamethoxazde (SXT) ≥ 4/75 0.25/4.75 Table 2. Putative antibiotic resistance genes and annotation of Cronobacter in this Study
Antibiotic Resistance Putative Gene Annotation Species C. malonaticus HB03 C. sakazaakii HB04 β-Lactam pbp1a
pbp1b
pbp2The enzyme has a penicillin-insensitive tranlycosylase N-terminal domain and a penicillin-insensitive transpeptidase C-teminal domain. +
+
++
+
+ampC
ampA, ampR
bl3_l
bl2be_
CTX-M-14
bl2be_shv2Class A, B, C β-lactamase. This enzyme breaks theβ-lactam antibiotic ring open and deactivates the molecule's antibacterial properties. +
+
+
-
++
-
+
+
-Aminoglycosides aac3iia Aminoglycoside N-acetyltransferase, which modifies aminoglycosides by acetylation. + - Macrolides aph3ia
aph33ib
aph6idAminoglycoside O-phosphotransferase, which modifies aminoglycosides by phosphorylation. +
+
+-
-
-Tetracycline ant2ia Aminoglycoside O-nucleotidylyltransferase, which modifies aminoglycosides by adenylylation. + - Phenicols erea Erythromycin esterase, which can inactivate erythromycin by lactone ring cleavage. + - tetd Major facilitator superfamily transporter, tetracycline efflux pump. + - tet34 Xanthine-guanine phosphoribosytransferase. + + cata2 Group A chloramphenicol acetyltransferase, which can inactivate chloramphenicol. + - Multidrug resistance efflux pump acra
acrb
emrd
emre
macb
mdtk
mdth
mdtg
oprm
tolc
ykkcMultidrug resistance efflux pump +
+
+
+
+
+
+
+
-
+
++
+
+
+
-
+
+
+
+
+
+ -
[1] Alsonosi A, Hariri S, Kajsík M, et al. The speciation and genotyping of Cronobacter isolates from hospitalised patient. Eur J Clin Microbiol Infect Dis, 2015; 34, 1979-88. doi: 10.1007/s10096-015-2440-8 [2] Gurtler JB, Kornacki JL, Beuchat LR. Enterobacter sakazakii:A coliform of increased concern to infant health. Int J Food Microbiol, 2005; 104, 1-34. doi: 10.1016/j.ijfoodmicro.2005.02.013 [3] Hariri S, Joseph S, Forsythe SJ. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg Infect Dis, 2013; 19, 175-7. doi: 10.3201/eid1901.120649 [4] Harris PNA, Ferguson JK. Antibiotic therapy for inducible AmpC β-lactamase-producing Gram-negative bacilli:what are the alternatives to carbapenems, quinolones and aminoglycosides? Int J Antimicrob Agentss, 2012; 40, 297-305. doi: 10.1016/j.ijantimicag.2012.06.004 [5] Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine, 2001; 80, 113-22. doi: 10.1097/00005792-200103000-00004 [6] Mshana SE, Gerwing L, Minde M, et al. Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania-International Journal of Antimicrobial Agents. Int J Antimicrob Agents, 2011; 38, 265-9. http://www.academia.edu/17624094/Outbreak_of_a_novel_Enterobacter_sp._carrying_blaCTX-M-15_in_a_neonatal_unit_of_a_tertiary_care_hospital_in_Tanzania [7] Müller A, Hächler H, Stephan R, et al. Presence of AmpC Beta-Lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 Conferring an Unusual Resistance Phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb Drug Resist, 2014; 20, 275-80. doi: 10.1089/mdr.2013.0188 [8] Pan Z, Cu J, Lyuet G, et al. Isolation and molecular typing of Cronobacter spp. in commercial powdered infant formula and follow-up formula. Foodborne Pathog Dis, 2014; 11, 456-61. [9] Patrick ME, Mahon Barbara E, Greene Sharon A, et al. Incidence of Cronobacter spp. infections, United States, 2003-2009. Emerg Infect Dis, 2014; 20, 1520-3. [10] Singh N, Goel G, Raghav M. Prevalence and Characterization of Cronobacter spp. fromVarious Foods, Medicinal Plants, and Environmental Samples. Curr Microbiol, 2015; 71, 31-8. doi: 10.1007/s00284-015-0816-8 -




 下载:
下载:



 Quick Links
Quick Links