-
Leclercia adecarboxylata is a motile, facultatively anaerobic, Gram-negative bacterium first identified by Leclerc in 1962[1]. L. adecarboxylata has been isolated from water, food, and other environmental sources, and is now recognized as a pathogenic organism[2]. Previous studies have reported the presence of this bacterium in immunocompromised patients; however, several recent cases of L. adecarboxylata have been reported in immunocompetent patients, particularly children[3-5].
Carbapenems are first-line antibiotics used to treat multidrug-resistant Gram-negative bacterial infections. However, carbapenem-resistant Enterobacteriaceae have become a major public health threat, leading to severe infections, limited treatment options, and mortality rates of 26%–44%. The prevalence of carbapenem resistance in Enterobacteriaceae is mediated by the rapid emergence of carbapenemase genes[6,7]. Although most cases of L. adecarboxylata infection are susceptible to common antibiotics, some drug-resistant strains have recently been detected[8-10]. A review of cases of L. adecarboxylata infection in humans revealed 82 publications describing clinical cases of L. adecarboxylata infection in 148 patients (104 adults and 44 children) since the first report in 1991[11-14]. Among the documented cases of pediatric infection, one case occurred in 1991, two in 2000–2004, six in 2011–2014, four in 2015–2019, and 31 in 2020–2022. L. adecarboxylata infection remains relatively rare; however, the number of reported cases in children has recently increased[15]. Here, we present a case of a carbapenem-resistant L. adecarboxylata strain in a newborn female to increase awareness of L. adecarboxylata as an emerging infection in children.
L. adecarboxylata 17YN198 was isolated in 2017 from the feces of a healthy 5-day-old female newborn with no relevant medical history in Yunnan, China. The strain was preliminarily identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Billerica, MA, USA)[16]. Identification was confirmed by 16S rRNA gene sequencing. Antimicrobial susceptibility was determined using the broth microdilution method, according to the Clinical Laboratory Standards Institute (CLSI) guidelines[17]. Twenty-nine common antimicrobial agents were used to evaluate antimicrobial susceptibility. The isolated strain exhibited resistance to ceftriaxone, ceftazidime, cefazolin, amoxicillin-clavulanate, trimethoprim-sulfamethoxazole, ertapenem, and meropenem (Table 1) according to the CLSI guidelines, and was susceptible to amikacin, gentamicin, aztreonam, chloramphenicol, norfloxacin, piperacillin-tazobactam, and minocycline.
Table 1. The MIC profile of 29 common antimicrobial agents for Leclercia adecarboxylata strain 17YN198
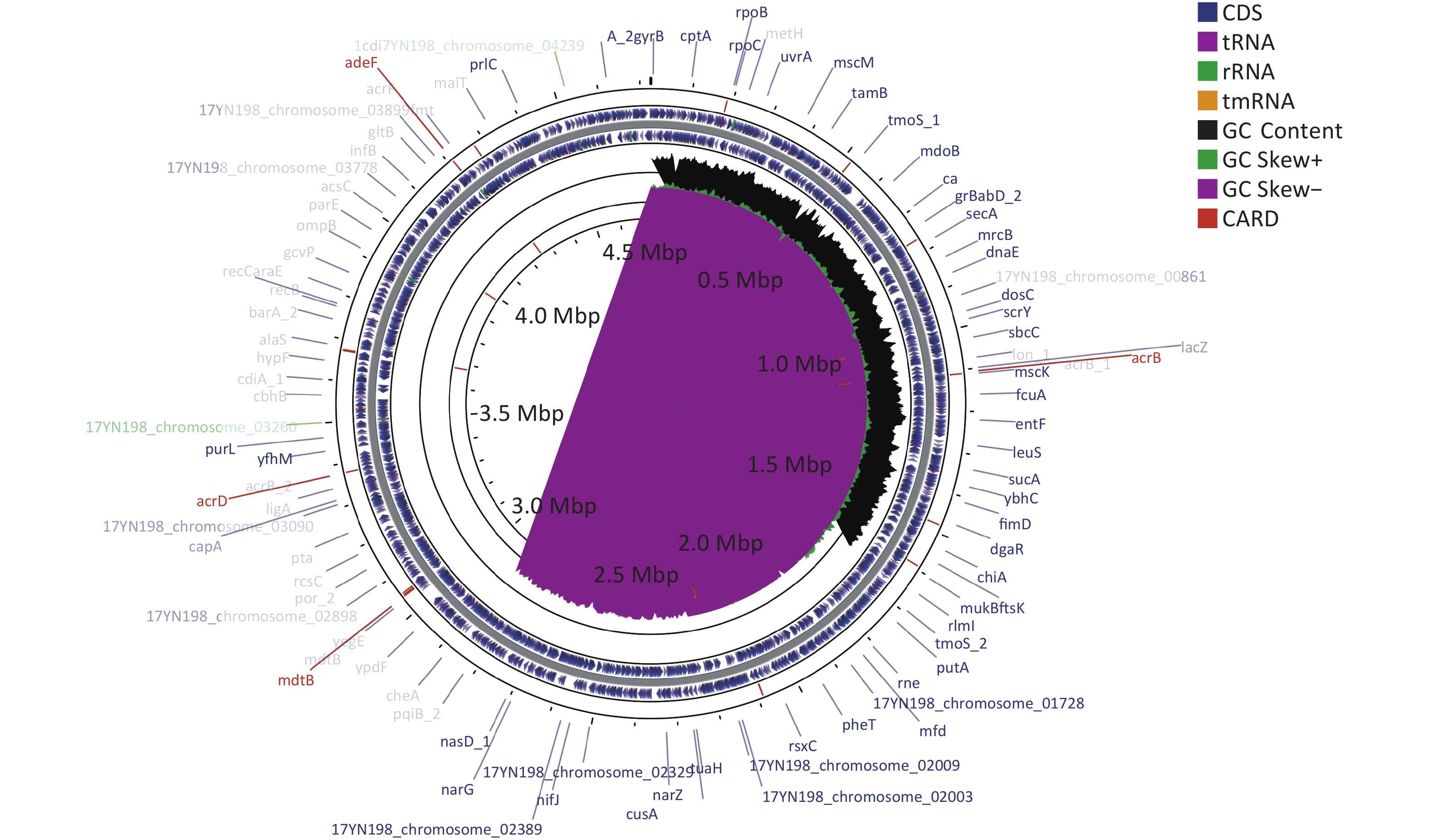
Drug classes Antibiotics MIC (µg/mL) Aminoglycosides Amikacin ≤ 8 Aminoglycosides Gentamicin ≤ 2 Aminoglycosides Tobramycin 8 β-lactam combination agents Amoxicillin – clavulanate 32/16 β-lactam combination agents Amoxicillin – sulbactam > 16/8 β-lactam combination agents Piperacillin – tazobactam ≤ 4/4 Monobactams Aztreonam ≤ 2 Cephems Cefazolin > 16 Cephems Cefepime 4 Cephems Cefoperazone – sulbactam > 32/8 Cephems Cefoxitin > 16 Cephems Ceftazidime > 32 Cephems Ceftriaxone 16 Cephems Cefuroxime > 16 Phenicols Chloramphenicol ≤ 4 Quinolones Ciprofloxacin ≤ 0.5 Quinolones Levofloxacin ≤ 1 Quinolones Moxifloxacin ≤ 0.5 Quinolones Norfloxacin ≤ 2 Lipopeptides Colistin 2 Carbapenems Ertapenem > 2 Carbapenems Imipenem 1 Carbapenems Meropenem 4 Fosfomycins Fosfomycin/G6P 128 Tetracyclines Minocycline 2 Tetracyclines Tetracycline > 8 Tetracyclines Tigecycline ≤ 1 Nitrofurans Macrodantin 32 Folate pathway antagonists Trimethoprim – sulfamethoxazole > 4/76 Note. MIC, minimum inhibitory concentration. Whole-genome sequencing was performed using the Illumina NovaSeq PE150 platform (Illumina, https://www.illumina.com) and PacBio single-molecule real-time sequencing[18]. Resistance genes were analyzed using ResFinder 2.1, and mobile elements were determined using bioinformatics tools provided by IS Finder. The entire L. adecarboxylata 17YN198 genome sequence was deposited in the GenBank database under the accession number CP106959-CP106963. L. adecarboxylata 17YN198 contained a single circular chromosome with a length of 4,725,550 base pairs (bp) and a GC content of 55.69%. The chromosome carried 4,305 protein-coding genes, 86 tRNA genes, and 25 rRNA genes (Figure 1).
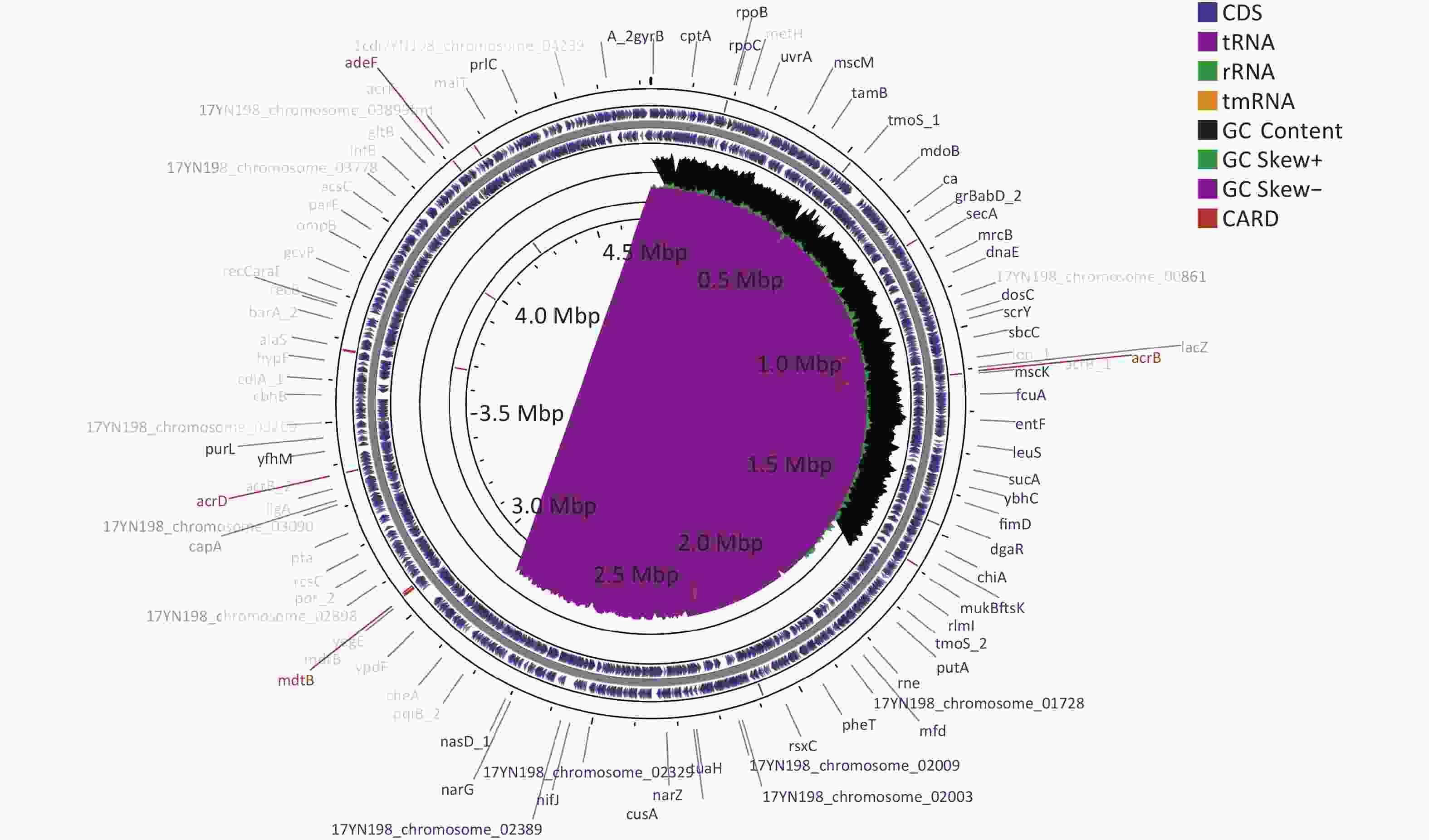
Figure 1. Circular map of the L. adecarboxylata 17YN198 genome was designed using CGView. The gene marked in red in the outermost circle is the drug resistance gene obtained from card database comparison. The outer ring denotes the ORFs on the positive strand. The next ring illustrates ORFs on the complementary strand. The black circle presents GC content. The inner rings show G+C content and G+C skew, where peaks represent the positive (outward) and negative (inward) deviation from the mean G+C content and G+C skew, respectively.
Analysis of acquired resistance genes using ResFinder 2.1 showed that L. adecarboxylata 17YN198 harbored 33 antimicrobial resistance genes encoding resistance to tetracyclines (tetA and tetR), carbapenems (blaIMP), aminoglycosides [aph(3')-Ia, aadA6, (AGly)aacA4, aadA1], fluoroquinolones (qnrB5), and folate pathway antagonists (sul1) (Table 2). According to the assembly results, the L. adecarboxylata 17YN198 isolate carried four plasmids. These included the 42,504-bp sul1-bearing plasmid pIMP-1 (CP106960), 155,030-bp plasmid 2 (CP106961), 115,001-bp blaIMP-harboring plasmid 3 (CP106962), and 52,474-bp plasmid 4 (CP106963). Plasmids 2 and 4 did not harbor any antimicrobial resistance genes. The results showed a susceptibility pattern consistent with the presence of blaIMP and sul1, that is, resistance to meropenem, ertapenem, and trimethoprim-sulfamethoxazole. The blaIMP gene harbored in the strain was most similar to blaIMP-79 at the nucleotide level, with a similarity of 99.40% (737/741). The carbapenemase IMP encoded by the blaIMP gene had 100% amino acid identity with carbapenemase IMP-1.
Table 2. Thirty-three resistance genes harbored in the L. adecarboxylata 17YN198
Chromosome Start End ARGs 17YN198_chromosome_1 1009446 1010621 (Bla)ampH 17YN198_chromosome_1 1104447 1107587 acrB 17YN198_chromosome_1 1107610 1108803 Enterobacter_cloacae_acrA 17YN198_chromosome_1 1177961 1178302 ramA 17YN198_chromosome_1 1271921 1273822 (Bla)Penicillin_Binding_Protein_Ecoli 17YN198_chromosome_1 1480168 1481403 mdf(A) 17YN198_chromosome_1 1591372 1593120 msbA 17YN198_chromosome_1 1784331 1785539 mdtH 17YN198_chromosome_1 2191719 2192102 marA 17YN198_chromosome_1 2680813 2681226 H-NS 17YN198_chromosome_1 2968917 2970359 mdtK 17YN198_chromosome_1 3042283 3043515 mdtA 17YN198_chromosome_1 3043515 3046640 mdtB 17YN198_chromosome_1 3051130 3052533 baeS 17YN198_chromosome_1 3052530 3053255 baeR 17YN198_chromosome_1 3173857 3175500 yojI 17YN198_chromosome_1 3375652 3378765 acrD 17YN198_chromosome_1 3671973 3672503 emrR 17YN198_chromosome_1 3672629 3673804 Klebsiella_pneumoniae_KpnG 17YN198_chromosome_1 3673821 3675368 emrB 17YN198_chromosome_1 3987117 3987935 bacA 17YN198_chromosome_1 4274088 4274720 CRP 17YN198_chromosome_1 4587843 4589216 cpxA 17YN198_plasmid1_1 70246 71445 (Tet)tetA 17YN198_plasmid1_1 71524 72201 (Tet)tetR 17YN198_plasmid1_1 80428 81243 aph(3')-Ia 17YN198_plasmid1_1 83245 83925 QnrB5 17YN198_plasmid1_1 86408 87247 sul1 17YN198_plasmid1_1 87752 88543 aadA1 17YN198_plasmid1_1 88636 89109 dfrA1 17YN198_plasmid3_1 30895 31740 aadA6 17YN198_plasmid3_1 31810 32307 (AGly)aacA4 17YN198_plasmid3_1 32826 33566 blaIMP-79 The blaIMP-harboring plasmid, designated as plasmid 3, had a length of 42,504 bp and an average GC content of 51.70%. Plasmid 3 belonged to the IncN3-incompatible group and contained three resistance genes [blaIMP-1, aadA6, and (AGly)aacA4] (Figure 2). Plasmid comparison using BLASTn and the NCBI plasmid database (ftp.ncbi.nlm.nih.gov:/refseq/release/plasmid/) revealed that plasmid 3 is a novel plasmid. Although plasmid 3 did not carry ISs elements, it carried a type IV secretion system (virB1, virB3–virB6, and virB8–virB11). This suggests that the transfer of resistance genes may be related to T4SS binding. Since L. adecarboxylata 17YN198 was isolated from a newborn female, blaIMP-1-harboring plasmid 3 may represent a newly emerging risk factor for the spread of carbapenemase resistance.
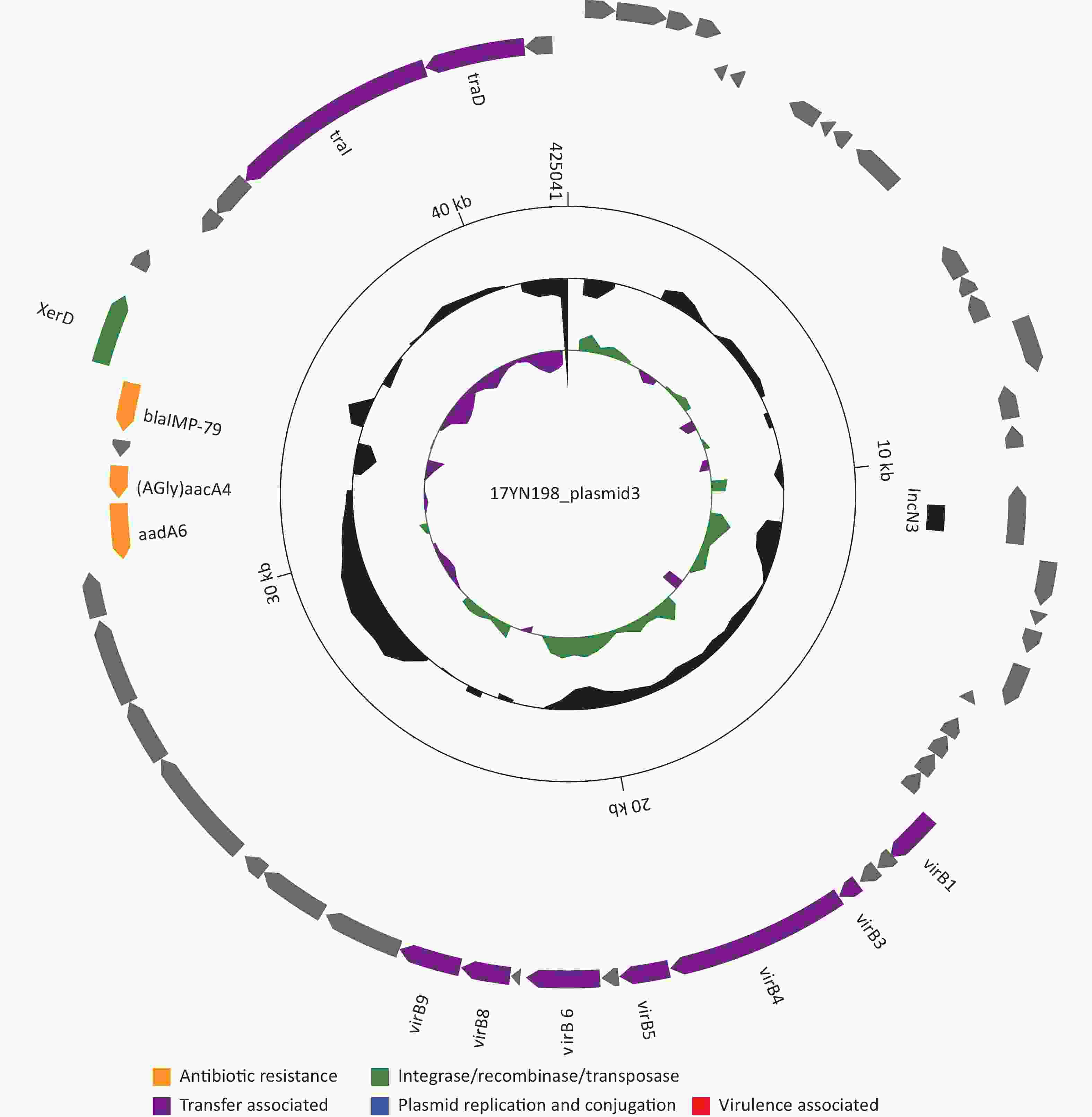
Figure 2. Schematic map of plasmid 3. Genes are denoted by arrows and colored based on gene function classification. The innermost circle presents GC-Skew (G-C/G+C) with a window size of 1,000 and step size of 500. The black circle presents GC content. Backbone and accessory module regions are also shown.
Infections caused by L. adecarboxylata have likely been underestimated for several decades because of the difficulty in correctly identifying the bacterium, leading to underreporting in the medical literature[19]. L. adecarboxylata was previously considered an opportunistic pathogen. However, this may be due to misdiagnosis because this organism shares several biochemical characteristics with Escherichia coli. L. adecarboxylata is now considered a pathogenic organism due, in part, to the use of modern identification techniques such as API 20E (bioMérieux, Craponne, France) and MALDI-TOF MS, which have been used to accurately identify and isolate L. adecarboxylata from E. coli.
To the best of our knowledge, this is the first report of carbapenem-resistant L. adecarboxylata isolated from a healthy newborn. The L. adecarboxylata strain isolated in this study carried four plasmids that may serve as reservoirs for antibiotic resistance genes. This case highlights the importance of considering L. adecarboxylata as a potential cause of infections in children. These findings suggest that close monitoring of resistant strains in the human gut microbiota should become routine clinical practice to prevent the occurrence of infections.
-
The datasets generated in this study are available in GenBank: SAMN31079621 and CP106959-CP106963.
doi: 10.3967/bes2023.104
-
Abstract: Leclercia adecarboxylata is a Gram-negative bacterium belonging to the Enterobacteriaceae family. To our knowledge, this is the first report of a carbapenem-resistant L. adecarboxylata strain isolated from a healthy newborn. The L. adecarboxylata strain isolated in this study carried four plasmids that may serve as reservoirs for antibiotic resistance genes. Plasmids 2 and 4 did not harbor any antimicrobial resistance genes. Plasmid 3 is a novel plasmid containing three resistance genes. The blaIMP gene harbored in the strain was most similar to blaIMP-79 at the nucleotide level, with a similarity of 99.4% (737/741). This case highlights the importance of considering L. adecarboxylata as a potential cause of infections in children.
-
Key words:
- Leclercia adecarboxylata /
- Carbapenem-resistant /
- Newborn
-
Figure 1. Circular map of the L. adecarboxylata 17YN198 genome was designed using CGView. The gene marked in red in the outermost circle is the drug resistance gene obtained from card database comparison. The outer ring denotes the ORFs on the positive strand. The next ring illustrates ORFs on the complementary strand. The black circle presents GC content. The inner rings show G+C content and G+C skew, where peaks represent the positive (outward) and negative (inward) deviation from the mean G+C content and G+C skew, respectively.
Figure 2. Schematic map of plasmid 3. Genes are denoted by arrows and colored based on gene function classification. The innermost circle presents GC-Skew (G-C/G+C) with a window size of 1,000 and step size of 500. The black circle presents GC content. Backbone and accessory module regions are also shown.
Table 1. The MIC profile of 29 common antimicrobial agents for Leclercia adecarboxylata strain 17YN198
Drug classes Antibiotics MIC (µg/mL) Aminoglycosides Amikacin ≤ 8 Aminoglycosides Gentamicin ≤ 2 Aminoglycosides Tobramycin 8 β-lactam combination agents Amoxicillin – clavulanate 32/16 β-lactam combination agents Amoxicillin – sulbactam > 16/8 β-lactam combination agents Piperacillin – tazobactam ≤ 4/4 Monobactams Aztreonam ≤ 2 Cephems Cefazolin > 16 Cephems Cefepime 4 Cephems Cefoperazone – sulbactam > 32/8 Cephems Cefoxitin > 16 Cephems Ceftazidime > 32 Cephems Ceftriaxone 16 Cephems Cefuroxime > 16 Phenicols Chloramphenicol ≤ 4 Quinolones Ciprofloxacin ≤ 0.5 Quinolones Levofloxacin ≤ 1 Quinolones Moxifloxacin ≤ 0.5 Quinolones Norfloxacin ≤ 2 Lipopeptides Colistin 2 Carbapenems Ertapenem > 2 Carbapenems Imipenem 1 Carbapenems Meropenem 4 Fosfomycins Fosfomycin/G6P 128 Tetracyclines Minocycline 2 Tetracyclines Tetracycline > 8 Tetracyclines Tigecycline ≤ 1 Nitrofurans Macrodantin 32 Folate pathway antagonists Trimethoprim – sulfamethoxazole > 4/76 Note. MIC, minimum inhibitory concentration. Table 2. Thirty-three resistance genes harbored in the L. adecarboxylata 17YN198
Chromosome Start End ARGs 17YN198_chromosome_1 1009446 1010621 (Bla)ampH 17YN198_chromosome_1 1104447 1107587 acrB 17YN198_chromosome_1 1107610 1108803 Enterobacter_cloacae_acrA 17YN198_chromosome_1 1177961 1178302 ramA 17YN198_chromosome_1 1271921 1273822 (Bla)Penicillin_Binding_Protein_Ecoli 17YN198_chromosome_1 1480168 1481403 mdf(A) 17YN198_chromosome_1 1591372 1593120 msbA 17YN198_chromosome_1 1784331 1785539 mdtH 17YN198_chromosome_1 2191719 2192102 marA 17YN198_chromosome_1 2680813 2681226 H-NS 17YN198_chromosome_1 2968917 2970359 mdtK 17YN198_chromosome_1 3042283 3043515 mdtA 17YN198_chromosome_1 3043515 3046640 mdtB 17YN198_chromosome_1 3051130 3052533 baeS 17YN198_chromosome_1 3052530 3053255 baeR 17YN198_chromosome_1 3173857 3175500 yojI 17YN198_chromosome_1 3375652 3378765 acrD 17YN198_chromosome_1 3671973 3672503 emrR 17YN198_chromosome_1 3672629 3673804 Klebsiella_pneumoniae_KpnG 17YN198_chromosome_1 3673821 3675368 emrB 17YN198_chromosome_1 3987117 3987935 bacA 17YN198_chromosome_1 4274088 4274720 CRP 17YN198_chromosome_1 4587843 4589216 cpxA 17YN198_plasmid1_1 70246 71445 (Tet)tetA 17YN198_plasmid1_1 71524 72201 (Tet)tetR 17YN198_plasmid1_1 80428 81243 aph(3')-Ia 17YN198_plasmid1_1 83245 83925 QnrB5 17YN198_plasmid1_1 86408 87247 sul1 17YN198_plasmid1_1 87752 88543 aadA1 17YN198_plasmid1_1 88636 89109 dfrA1 17YN198_plasmid3_1 30895 31740 aadA6 17YN198_plasmid3_1 31810 32307 (AGly)aacA4 17YN198_plasmid3_1 32826 33566 blaIMP-79 -
[1] Leclerc H. Biochemical study of pigmented Enterobacteriaceae. Ann Inst Pasteur (Paris), 1962; 102, 726−41. [2] Keren Y, Keshet D, Eidelman M, et al. Is Leclercia adecarboxylata a new and unfamiliar marine pathogen?. J Clin Microbiol, 2014; 52, 1775−6. doi: 10.1128/JCM.03239-13 [3] Grantham WJ, Funk SS, Schoenecker JG. Leclercia adecarboxylata musculoskeletal infection in an immune competent pediatric patient: An emerging pathogen?. Case Rep Orthop, 2015; 2015, 160473. [4] Keyes J, Johnson EP, Epelman M, et al. Leclercia adecarboxylata: An emerging pathogen among pediatric infections. Cureus, 2020; 12, e8049. [5] Hurley EH, Cohen E, Katarincic JA, et al. Leclercia adecarboxylata infection in an immunocompetent child. R I Med J (2013), 2015; 98, 41−4. [6] Haji SH, Aka STH, Ali FA. Prevalence and characterisation of carbapenemase encoding genes in multidrug-resistant Gram-negative bacilli. PLoS One, 2021; 16, e0259005. doi: 10.1371/journal.pone.0259005 [7] Awoke T, Teka B, Aseffa A, et al. Detection of bla KPC and bla NDM carbapenemase genes among Klebsiella pneumoniae isolates in Addis Ababa, Ethiopia: Dominance of bla NDM . PLoS One, 2022; 17, e0267657. doi: 10.1371/journal.pone.0267657 [8] Shin GW, You MJ, Lee HS, et al. Catheter-related bacteremia caused by multidrug-resistant Leclercia adecarboxylata in a patient with breast cancer. J Clin Microbiol, 2012; 50, 3129−32. doi: 10.1128/JCM.00948-12 [9] Garza-González E, Bocanegra-Ibarias P, Rodríguez-Noriega E, et al. Molecular investigation of an outbreak associated with total parenteral nutrition contaminated with NDM-producing Leclercia adecarboxylata. BMC Infect Dis, 2021; 21, 235. doi: 10.1186/s12879-021-05923-0 [10] Alosaimi RS, Muhmmed Kaaki M. Catheter-related ESBL-producing Leclercia adecarboxylata septicemia in hemodialysis patient: An emerging pathogen?. Case Rep Infect Dis, 2020; 2020, 7403152. [11] Spiegelhauer MR, Andersen PF, Frandsen TH, et al. Leclercia adecarboxylata: a case report and literature review of 74 cases demonstrating its pathogenicity in immunocompromised patients. Infect Dis (Lond), 2019; 51, 179−88. doi: 10.1080/23744235.2018.1536830 [12] Broderick A, Lowe E, Xiao A, et al. Leclercia adecarboxylata folliculitis in a healthy swimmer-An emerging aquatic pathogen?. JAAD Case Rep, 2019; 5, 706−8. doi: 10.1016/j.jdcr.2019.06.007 [13] Adapa S, Konala VM, Javed T, et al. Peritonitis from Leclercia adecarboxylata: An emerging pathogen. Clin Case Rep, 2019; 7, 829−31. doi: 10.1002/ccr3.2094 [14] Sun Q L, Wang HY, Shu LB, et al. Leclercia adecarboxylata from human gut flora carries mcr-4.3 and bla IMP-4 -bearing plasmids. Front Microbiol, 2019; 10, 2805. doi: 10.3389/fmicb.2019.02805 [15] Hassan I, Gupta P, Ray P, et al. Leclercia adecarboxylata causing spontaneous bacterial peritonitis in a child with nephrotic syndrome: A case report and review of literature. J Lab Physicians, 2020; 12, 222−4. doi: 10.1055/s-0040-1721162 [16] Lévesque S, Dufresne PJ, Soualhine H, et al. A side by side comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One, 2015; 10, e0144878. doi: 10.1371/journal.pone.0144878 [17] Gajic I, Kabic J, Kekic D, et al. Antimicrobial susceptibility testing: A comprehensive review of currently used methods. Antibiotics (Basel), 2022; 11, 427 . doi: 10.3390/antibiotics11040427 [18] Li RC, Xie MM, Dong N, et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. GigaScience, 2018; 7, 1−9. [19] Aarab A, Saddari A, Noussaiba B, et al. Leclercia adecarboxylata invasive infection in a patient with Hirschsprung disease: A case report. Ann Med Surg (Lond), 2021; 71, 102927. doi: 10.1016/j.amsu.2021.102927 -




 下载:
下载:



 Quick Links
Quick Links