-
Silica has been recognized as a hazardous material for centuries[1]. Inhalation of crystalline silica particles, particularly in occupational settings such as mining, construction, and agriculture, can lead to severe respiratory complications and long-term health consequences, the most prominent being silicosis. It manifests as persistent inflammation and diffuse fibrosis of the lung interstitium. Despite the implementation of active prevention and control measures worldwide the number of individuals with a history of occupational dust exposure and newly reported cases of silicosis continues to grow[2,3]. Additionally, the pathogenesis of silicosis remains unclear, resulting in a lack of effective clinical interventions. Therefore, it is imperative to elucidate the underlying mechanisms of silica-induced lung inflammation and fibrosis.
Recent studies have demonstrated that lung microbiota dysbiosis is associated with the pathogenic mechanisms of various lung diseases, such as chronic obstructive pulmonary disease (COPD), bronchial asthma, tuberculosis, and pneumonia. These studies have also highlighted that the critical role of pulmonary commensal bacteria in regulating the homeostasis and function of the host immune system[4-6]. Silicosis, a heterogeneous disease, is characterized by the uncontrolled deposition of collagen in the lung interstitium, accompanied by the thickening of the alveolar-capillary membrane, which gradually impairs gas exchange[7]. These anatomical, physiological, and immunological characteristics of pulmonary fibrosis can exert selective pressure on the bacterial communities in the lungs, leading to microbiota dysbiosis[4]. Yang et al. were the first to confirm that lung microbiota dysbiosis can drive the occurrence and progression of bleomycin-induced pulmonary fibrosis. The findings revealed an altered microbial composition in the lungs of bleomycin-exposed mice compared to that of the control group. However, mice with antibiotic-induced microbiota depletion or germ-free mice exhibited significantly reduced immune cell infiltration and pulmonary fibrotic responses, demonstrating effective resistance to bleomycin-induced lethal lesions[8]. A recent experimental study suggested that dysbiosis of the lung microbiota in mice enhances silica-induced fibrosis through the lipopolysaccharide/Toll-like receptor 4 pathway[9]. These findings indicate that the pulmonary microbiota is closely associated with pulmonary fibrosis and suggest that pulmonary commensal bacteria may play a key role in alleviating this condition. However, the current issue revolves around revealing the characteristics of commensal bacteria in patients with silicosis or animal models, and studies on how dysbiotic microbiota affect development of silicosis remain limited. In particular, other major classes of biomolecules that constitute mucosal homeostasis, such as surfactant proteins (SPs), have been overlooked in research on pulmonary commensal bacteria.
SPs, including SP-A, SP-B, SP-C, and SP-D, have been extensively studied for their roles in pulmonary surfactant function and immune regulation[10]. Notably, SP-A and SP-D, belonging to the collection protein family, exhibit antimicrobial properties and participate in the recognition and clearance of pathogens, modulation of inflammatory responses, and maintenance of mucosal immunity. SP-B and SP-C are primarily responsible for reducing surface tension, preventing alveolar collapse, facilitating gas exchange, and maintaining lung compliance by regulating the surfactant spread, phospholipid recycling, cell membrane permeability, and transmembrane action[11,12]. Recent studies have shown that SPs can directly interact with certain commensal bacteria, influencing their growth, colonization, and virulence. Additionally SPs can indirectly modulate the composition and diversity of the lung microbiota through their effects on the host immune system and epithelial barrier integrity. These findings suggest that the abnormal expression of SPs and disorders of pulmonary commensal bacteria may play a crucial role in various lung diseases, including silicosis[13-15]. However, the roles of lung microbiota and SPs in silicosis remain unclear. Thus, the relationship between lung microbiota and SPs in silicosis requires further exploration.
This study explored the association between the abnormal expression of SPs and silicosis by observing dynamic changes in SPs levels in lung tissues. Additionally, we investigated changes in lung microbiota in a silicosis mouse model using 16S rRNA gene sequencing analysis. Furthermore, based on Abx mice, we examined the relationship between lung microbiota dysbiosis and the abnormal expression of SPs during silica-induced lung fibrosis. These results provide experimental evidence to further elucidate the pathogenesis of silicosis and underscore the importance of maintaining pulmonary mucosal homeostasis during its treatment.
-
The silicosis mouse model was generated using SiO2-induced (–99% SiO2, 3–5 µm particle size, SILICA, USA). Briefly, SiO2 was ground in an agate mortar for 2 h and then placed in a beaker and heated in a pyrostat at 180 °C for 2 h to dehydrate and remove endotoxins. subsequently, the silica particulates were suspended in sterile saline (50 mg/mL), followed by sonication for 10 min and vortexed for 3 s before instillation.
Three-week-old male C57BL/6N mice were purchased from the Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) and housed in a specific pathogen-free (SPF) environment. The mice were maintained on standard chow at a temperature of 24 ± 1 °C with a 12/12 h light/dark cycle with ad libitum access to water.
-
Intratracheal instillation of silica was used to establish a mouse silicotic model[16]. The experimental design and animal treatment protocols are illustrated in Figure 1A. Briefly, the mice were anesthetized and instilled with either 50 μL of Phosphate Buffer Saline (1X PBS) as the control group or 50 mg/mL silica suspension in PBS via the laryngotracheal route[17,18]. The mice were anesthetized using sodium pentobarbital (50 mg/kg) and sacrificed to harvest lung tissues on days 7, 14, 28, and 56 after intratracheal instillation (n = 5 mice per group).
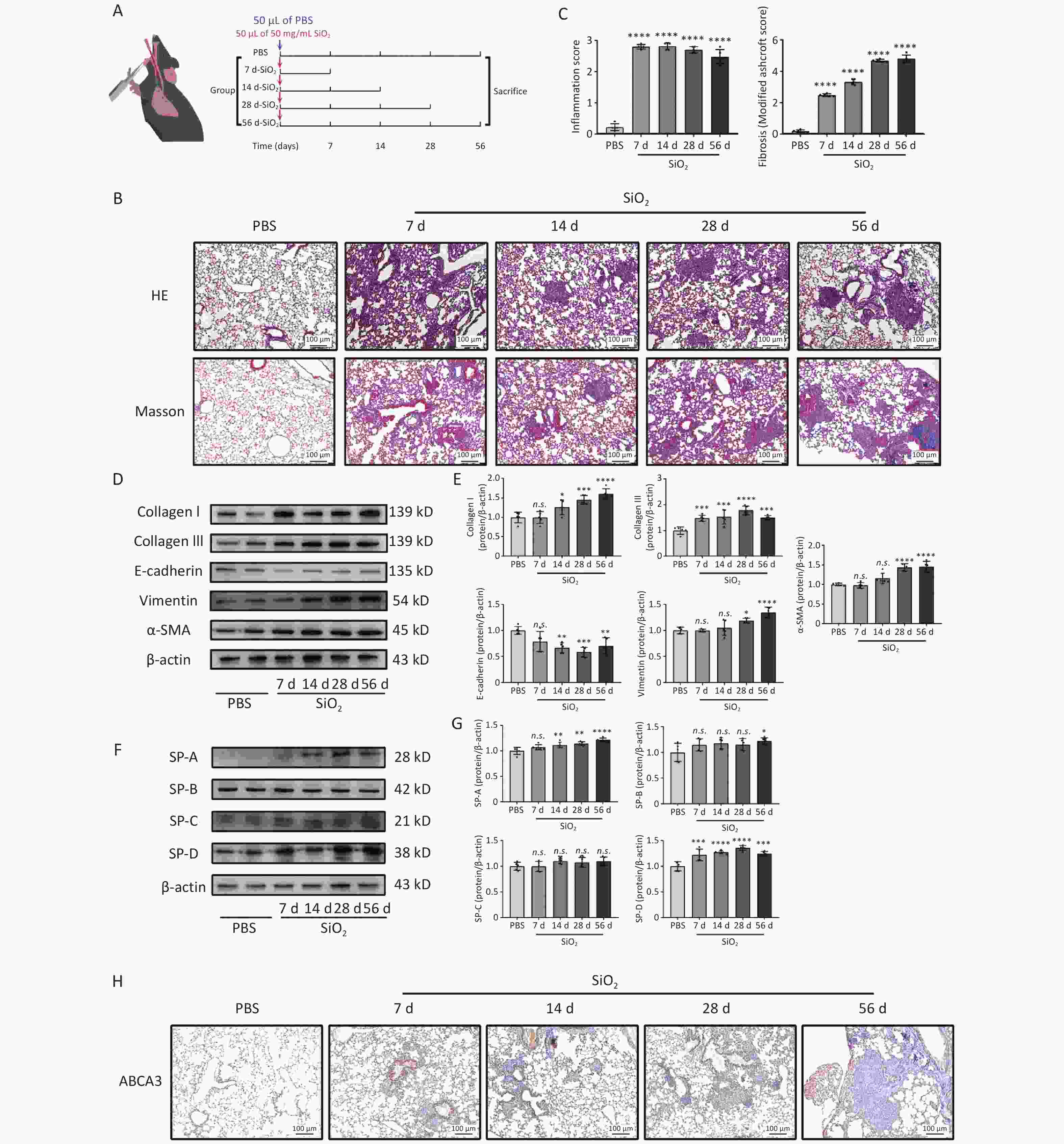
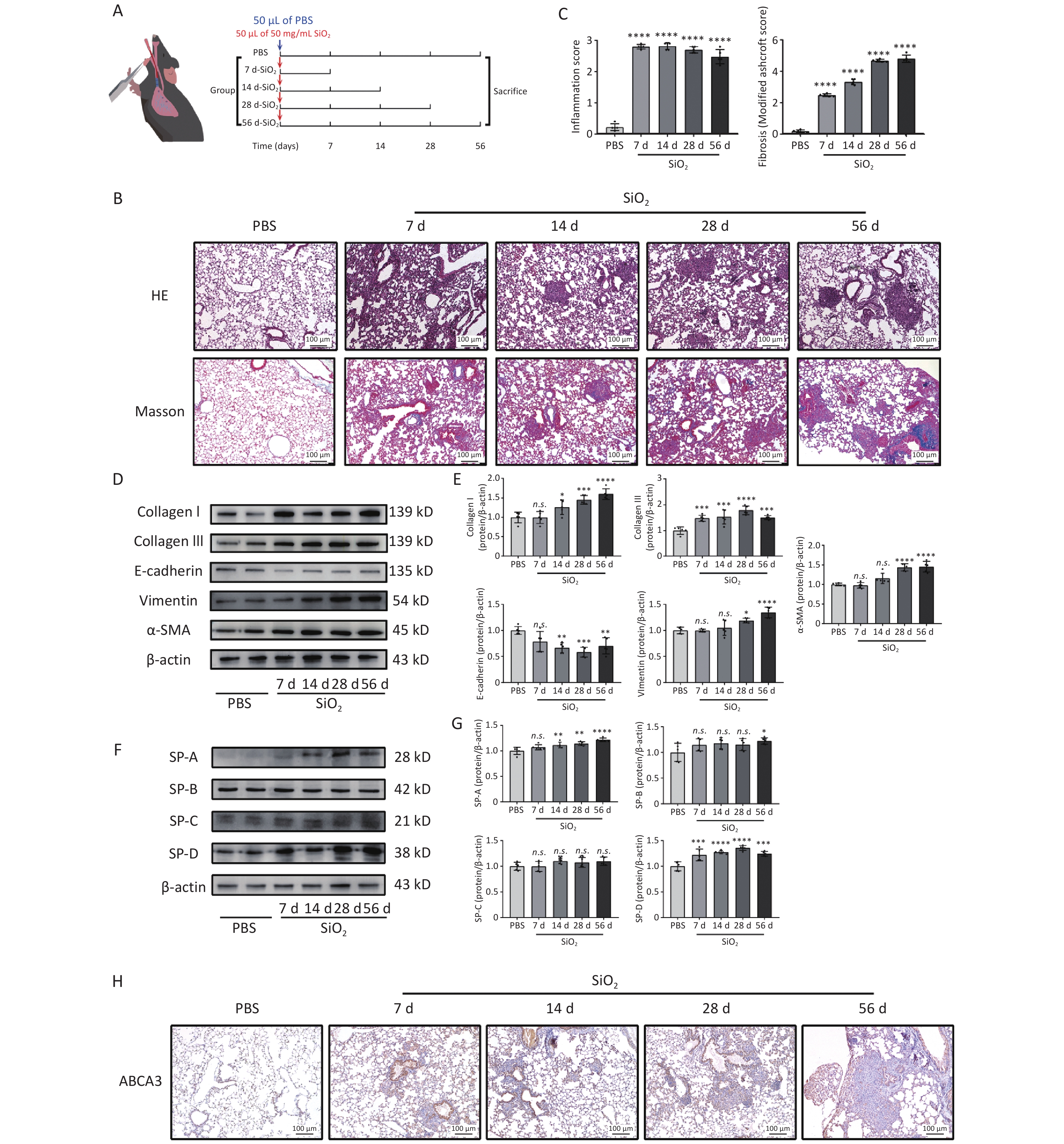
Figure 1. Effects of silica exposure on lung fibrosis and surfactant proteins (SPs). (A) Treatment protocol of the first subexperiment. (B) Representative photomicrographs of HE and Masson staining of lungs sections from mice. Scale bars: 100 μm. (C) Histologic semiquantitative scores (0–3) for inflammation and fibrosis as assessed by Modified Ashcroft Scale (0–8). (D) Expression levels of fibrosis marker proteins: Collagen I, Collagen III, E-cadherin, Vimentin, and α-SMA, in the lung tissue of mice in different groups, as measured by western blotting. (E) Corresponding densitometry data of Collagen I, Collagen III, E-cadherin, Vimentin, and α-SMA protein levels quantified using ImageJ software. (F) Representative images of SPs, SP-A, -B, -C, and -D, in the lung tissue from different groups. (G) Corresponding densitometry data of SP-A, -B, -C, and -D protein levels quantified using ImageJ software. Bar graphs are presented as the mean ± SD (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. presents no statistically significant difference. (H) Representative photomicrographs of ABCA3 immunohistochemical staining in the lung tissue from mice.
-
Mice in the antibiotics treatment (Abx) group were intranasally administered 50 μL of antibiotic cocktail containing ampicillin (1 mg/mL; Absin), vancomycin (0.5 mg/mL; Absin), metronidazole (1 mg/mL; Absin), and neomycin (1 mg/mL; Absin) once a week for 4 weeks. During this period, they also given the same antibiotic cocktail in their drinking water ad libitum for four consecutive weeks[8]. Mice in the vehicle group received sterilized water (n = 5 mice per group). Twenty-eight days after the first Abx treatment the mice were sacrificed to collect biological samples.
-
Mice pretreated with an antibiotic cocktail for 4 weeks or sterile water, were exposed to silica or sterile PBS. The mice were sacrificed 28 days after silica exposure (n = 5 mice per group).
-
After sacrificing the mice, the right lung was immediately tied off, and the left lung was injected with 1 mL of sterile PBS thrice to obtain bronchoalveolar lavage fluid (BALF). The right lower lung lobes were fixed in buffered formalin for histopathological evaluation.
The remaining lung tissues were cut into small pieces and stored at –80 °C for Western blot or qPCR analysis.
-
After fixation in 4% formalin for 48 h, the inferior lobe of the right lung was embedded in paraffin, and sections were cut to a thickness of 4 μm and subjected to hematoxylin and eosin (H&E), Masson tricolor (Masson), and immunohistochemistry staining, separately. Representative images of the four groups were acquired using an upright light microscope (Leica DM3000; Carl). Alveolar lung development was evaluated using the (Radial alveolar counts) RAC[19] and mean linear intercept (MLI)[20]. The level of inflammation in the lung tissues of mice was assessed based on the inflammatory cell counts in H&E-stained slides[21]. For the Masson-stained slides, the degree of pulmonary fibrosis was scored using the Modified Ashcroft Scale[22].
For immunohistochemical staining, sections of formalin-fixed paraffin-embedded lung tissue blocks were deparaffinized using xylene, followed by hydration through a graded series of ethanol solutions. Antigen retrieval was performed using a 10 mmol/L Tris/EDTA buffer solution (pH 9.0) at 100 °C for 20 min. After blocking, the sections were exposed to anti-ABCA3 antibody. Chromogenic detection was performed using a peroxidase-conjugated secondary antibody (30 min) and DAB reagent (5 min). Immunohistochemical images were captured using an upright light microscope (Leica DM3000; Carl).
-
Total DNA was extracted from the BALF samples using the FastPure Microbiome DNA Isolation Kit (Vazyme, cat#DC502), following the manufacturer’s protocol. The following primers were used for amplification: 16s rDNA forward primer 5’-GGTGAATACGTTCCCGG-3’ and reverse primer 5’-TACGGCTACCTTGTTACGACTT-3′. Then relative levels of the 16s rRNA gene were analyzed by SYBR GreenER™ qPCR SuperMix Universal (Invitrogen, USA) with LightCycler 96 (Roche, SE). The relative quantity of 16s rDNA in the same amount of total DNA was calculated using the Delta Ct method.
-
Total RNA was isolated from the upper left lung lobe of mice using the TRIzol™ Plus RNA Purification Kit (Invitrogen, USA), and then reverse transcribed into cDNA utilizing the SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, USA). The primer sequences are listed in Table 1. The cDNA was amplified using a real-time PCR detection system (Roche, SE). The housekeeping gene β-actin served as the internal control, and the relative mRNA expression levels were determined by 2−ΔΔCT values.
Table 1. Primer sequences used for qPCR
Gene Type Sequence Product (bp) TGF-β1 Forward 5’-TGGAGCAACATGTGGAACTC-3’ 70 Reverse 5’-CAGCAGCCGGTTACCAAG-3’ TNF-α Forward 5’-TCCCAGGTTCTCTTCAAGGGA-3’ 50 Reverse 5’-GGTGAGGAGCACGTAGTCGG-3’ IL-6 Forward 5’-TTCCATCCAGTTGCCTTCTTG-3’ 50 Reverse 5’-GAAGGCCGTGGTTGTCACC-3’ β-actin Forward 5’-TGTCCACCTTCCAGCAGATGT-3' 100 Reverse 5’-AGCTCAGTAACAGTCCGCCTAG-3' -
Protein extracts were separated using SDS-PAGE and transferred onto 0.45 μm polyvinylidine difluoride (PVDF) membranes. The following rabbit-derived primary antibodies were used: Collagen I (1:1,000), Collagen III (1:1,000), α-SMA (1:200) (Abcam, USA), E-cadherin (1:1,000) (Proteintech, USA), Vimentin (1:1,000) (Cell Signaling Technology Inc., USA), SP-A (1:1,000), SP-B (1:1,000), SP-C (1:1,000), SP-D (1:1,000), and β-Actin (1:3,000) (Affinity Biosciences, USA). The membranes were incubated with these antibodies overnight, washed thrice with 5% Tris-buffered saline Tween (TBST), and then incubated for 2 h with an HRP-conjugated goat anti-rabbit secondary antibody (1:3,000). Images were captured and analyzed using the FluorChem R multifunction imaging analysis system (ProteinSimple, California, USA) and ImageJ software.
-
The previously documented protocol detailed the perfusion fixation process using 1.5% paraformaldehyde (PFA) and 1.5% glutaraldehyde (GA) in a 0.15 mol/L HEPES buffer (pH 7.4). This procedure includes tissue sampling and the subsequent preparation of lung specimens for conventional transmission electron microscopy (TEM). Fixed lung tissue blocks, measuring 1 mm3, were carefully embedded in Agar 100 Resin® (Agar, Essex, England), an epoxy-based medium, and polymerized at 60 °C for 3 days. Embedded tissue blocks were then precisely trimmed using a diamond trimmer (Reichert TM 60, Austria). Subsequently, ultrathin sections (80 nm) were sliced using a Leica Ultracut E ultramicrotome (Leica, Nussloch, Germany). These sections were then treated with uranyl acetate for 2 min and lead citrate for 45 s to enhance contrast. The processed sections were carefully examined using a LEO 906 transmission electron microscope (LEO Electron Microscopy, Oberkochen, Germany) equipped with a 2k-camera (TRS, Troendle systems).
-
Microbial DNA was extracted from BALF samples using the FastPure Microbiome DNA Isolation Kit (Vazyme, cat#DC502). The 16S rRNA V3-V4 variable region was sequenced by Personal Biotechnology Co., Ltd. (Shanghai, China). Briefly, PCR was performed using the universal primer sequences 338 F (ACTCCTACGGGAGGCAGCA) with 7 bp barcodes (GAGTTCG) and 806R (GGACTACHVGGGTWTCTAAT) to amplify the V3–V4 region of the bacterial 16S rRNA gene. The resulting 16S rRNA amplicons were sequenced on Illumina MiSeq PE250 platform. After demultiplexing the raw sequence data using the primer sequences, quality filtering, trimming, and denoising were performed using the Divisive Amplicon Denoising Algorithm 2 method to acquire the amplicon sequence variant (ASV) table and annotate the species. QIIME2[23] was used to calculate α- and β-diversity and to analyze the species composition of the lung microbiota in mice. Taxonomic assignment of ASVs was performed using the feature-classifier plugin[24] against Greengenes 13_8. Additionally, based on the 16S rRNA gene sequencing data and Kyoto Encyclopedia of Genes and Genome (KEGG) orthology, PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) analysis and the Statistical Analysis of Metagenomic Profiles (STAMP) software package were used for the functional prediction of the lung microbiota.
-
Statistical analyses were performed using GraphPad Prism version 7.2.0 (GraphPad Software, Inc., California, USA). Data are expressed as the mean ± standard deviation (SD). Statistical comparisons among the four groups were performed using one-way ANOVA. Spearman’s rank correlation analysis was used for non-normally distributed data. For the corresponding multiple comparisons between individual groups, Tukey’s multiple comparison test was performed as a post-hoc test, and P < 0.05 represented statistical significance. LEfSe analysis was performed to estimate the effect sizes of the biomarkers, and the results are presented as LDA scores. A score > 2 and P < 0.05 indicated a marker species.
-
As shown in Figure 1B, on days 7 and 14 after following silica exposure, lung injuries in mice were mainly characterized by inflammatory cell infiltration, widened alveolar septum, and a thickened alveolar wall. By day 28 post-exposure, nodule formation in the cell fibers was evident. After 56 days of silica exposure, the lung tissue exhibited severe fibrosis, characterized by extensive extracellular matrix deposition and prominent collagen fibers, resulting in the marked disruption of normal lung architecture. Additionally, these pathological changes were accompanied by significantly elevated expression levels of collagen I, collagen III, α-SMA, and vimentin in lung tissues, compared with those in the control group (Figure 1D and F). The expression of E-cadherin in lung tissues significantly decreased after 14 days of silica exposure (Figure 1D and F). In response to silica stimulation, the expression levels of SP-A, SP-B, and SP-D increased compared to those in the PBS control group (Figure 1F and G). Immunohistochemical analysis revealed a significant rise in the number of ABCA3-positive ATII cells, that are responsible for storing and secreting surfactants, following silica stimulation. Moreover, a higher concentration of positive cells surrounding the cellular nodules. (Figure 1H). These findings confirm that ATII cells injury and abnormal SPs expression were associated with silica-induced pulmonary fibrosis.
-
To evaluate the effect of silica exposure on the pulmonary microbiota, we performed 16S rRNA analysis to examine the microbial community composition in the BALF of mice from both the PBS and SiO2 groups after a 28-day exposure period. A total of identified 1,334 ASVs were shared between the PBS and SiO2 groups (Figure 2A). Although statistical analysis using four indices (Chao1, observed_species, Shannon, and Simpson) revealed no significant differences in the alpha diversity of the lung microbiota between the control and SiO2 groups (Figure 2B, C, D, and E), the LEfSe analysis indicated a difference in the abundance of certain bacterial taxa between the two groups. Specifically, the abundance of certain taxa, such as the phylum Thermi and Actinobacteria, and the genera Acinetobacter and Brachybacterium, were significantly enriched in the silica-exposed group (Figure 2F). Principal Coordinates Analysis (PcoA) further demonstrated that silica exposure substantially altered the lung microbiota, explaining 36.9% (PCo1) and 19.3% (PCo2) of the observed variations in microbial composition (Figure 2G). These findings suggest that silica exposure drives dysbiosis of the pulmonary microbiota.
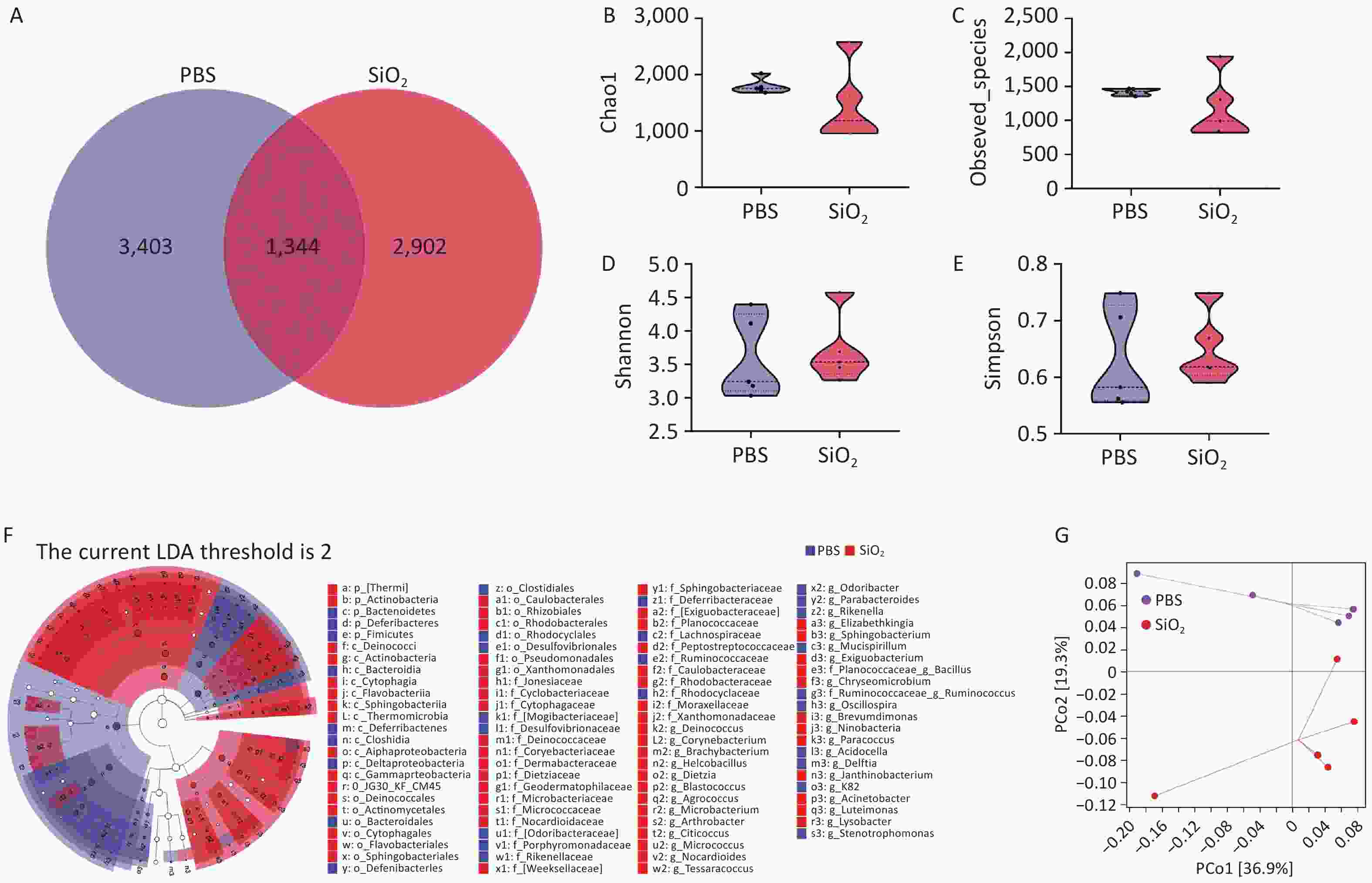
Figure 2. Overall evaluation effects of silica exposure on the lung microbiota using bronchoalveolar lavage fluid (BALF) samples. (A) Venn diagram analysis of the amplicon sequence variants (ASVs) shared between the control and silica groups. α-diversity estimators of lung bacterial community, including (B) Chao1, (C) observed species, (D) Shannon, and (E) Simpson. (F) LEfSe analysis to identifying the taxonomic biomarkers associated between silica exposure, where LDA scores indicate the strength of the association between identified taxa and the different treatments. (G) Principal Coordinates Analysis (PcoA) plots illustrating differences in the pulmonary microbiota from both groups.
-
We investigated the effects of commensal bacteria on the alveolar structure and expression of SPs in mice based on the Second Subexperiment (Figure 3A). These results demonstrated that Abx treatment significantly reduced the bacterial burden in the lungs (Figure 3B). Our findings revealed that mice treated with Abx exhibited alveolar structures that did not significantly differ from those of non-Abx-treated (vehicle) mice (Figure 3C and D). However, the expression levels of SP-A and SP-D were significantly increased in the lung tissues of Abx-treated mice (Figure 3E and F). Despite this, Abx-treated mice did not exhibit a statistically significant difference in the number of ABCA3-positive cells compared to non-Abx-treated mice (Figure 3G). These results suggested that Abx-induced depletion of the lung microbiota might disrupt the expression of pulmonary SPs, as evidenced by the increased expression of SP-A and SP-D in mice.
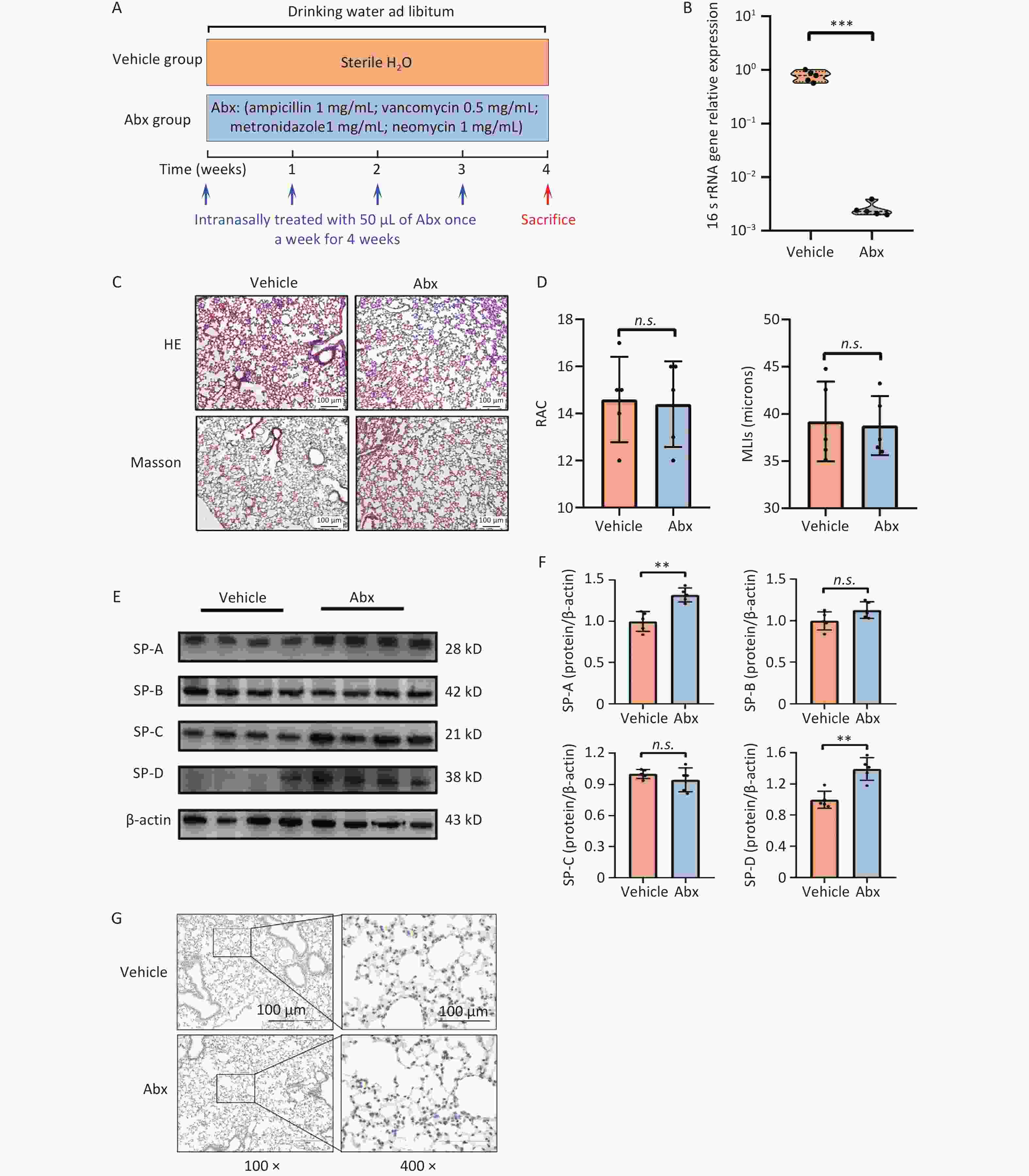
Figure 3. Effects of Abx-induced lung microbiota depletion on the lung physiology. (A) Treatment protocol of the second subexperiment in the study. (B) Expression levels of 16s rRNA gene. (C) Representative photomicrographs of hematoxylin-eosin (H & E) and Masson sections of lungs from mice. Scale bars: 100 μm. (D) Radial alveolar counts (RAC) and mean linear intercept (MLIs) of mice from the vehicle and Abx groups. (E) Expression levels of SP-A, -B, -C, and -D in the lung tissue, as measured by western blotting. (F) Corresponding densitometry data of SP-A, -B, -C, and -D protein levels were quantified using ImageJ software. Bar graphs are presented as the mean ± SD (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001, n.s. presents no statistically significant difference. (G) Representative photomicrographs of ABCA3 immunohistochemical staining in the lung tissue from different groups.
-
To investigate the effect of lung microbiota on silicosis pathogenesis, the third sub-experiment was conducted. (Figure 4A). The pathological results indicated that mice in the Abx+SiO2 group exhibited less lung inflammation and tissue damage than those in the SiO2 group (Figure 4B and C). Furthermore, the SiO2 group showed higher expression levels of collagen I, collagen III, vimentin, and α-SMA compared to those in both PBS group and Abx+SiO2 groups. While the SiO2 group also exhibited increased levels of collagen III and α-SMA compared to the Abx+SiO2 group, these differences did not reach statistical significance (Figure 4D & E). Additionally, the mRNA levels of TGF-β1, TNF-1α, and IL-6 were elevated in the lung tissues following silica exposure. However, these markers were reduced in the Abx+SiO2 group compared to that of the SiO2 group, and the difference of TGF-β1 was statistically significant (Figure 4F).
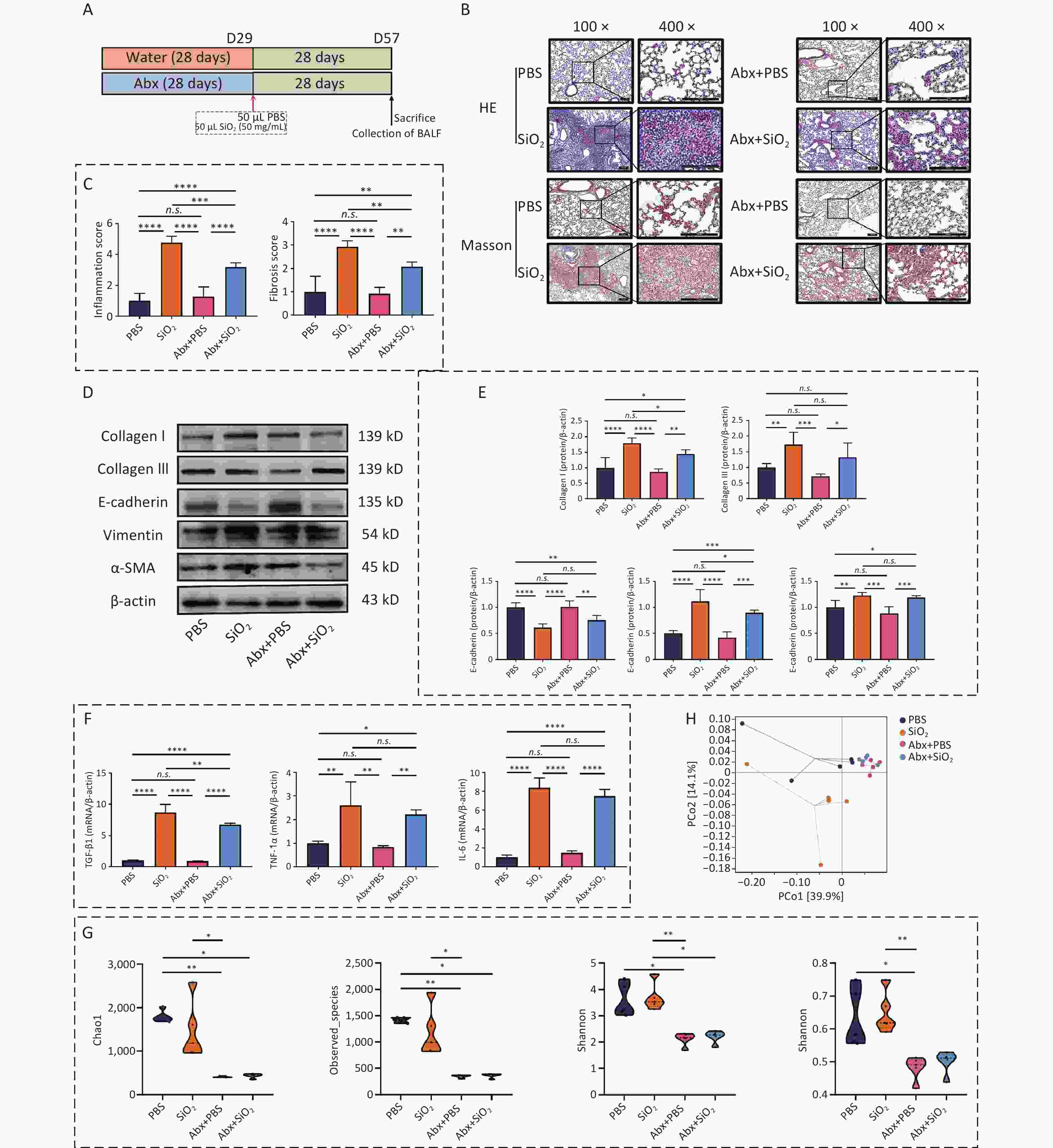
Figure 4. Combined antibiotic treatment mitigates silica-induced inflammatory pathology and fibrosis in pulmonary tissues by altering the microbiota. (A) Treatment protocol of the third subexperiment in the study. (B) Representative photomicrographs of HE and Masson sections of lungs from mice. Scale bars: 100 μm. (C) Histological semiquantitative score (0–3) of inflammation and the Modified Ashcroft Scale (0–8) for fibrosis. (D) Expression levels of Collagen I, Collagen III, E-cadherin, vimentin, and α-SMA in lung tissue, as measured by Western blotting. (E) Corresponding densitometry data for collagen I, collagen III, E-cadherin, vimentin, and α-SMA protein levels quantified using ImageJ Software. (F) qPCR analysis of the relative quantitative expression of TGF-β1, TNF-1α, and IL-6 in the pulmonary tissue, normalized to β-actin. Bar graphs are presented as the mean ± SD (n = 4 or 5). (G) α-diversity estimators of lung bacterial community. (H) PCoA plots showing differences in the pulmonary microbiota among groups. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. indicates no statistically significant difference.
Furthermore, the Shannon index indicated a statistically significant difference between the Abx+SiO2 and SiO2 groups (Figure 4G). The PcoA analysis also revealed a clear separation between the PBS and SiO2 groups, while the Abx+SiO2 group exhibited closer proximity to the control group. (Figure 4H). These findings indicated that silica exposure induced pathological changes associated with inflammatory and fibrotic responses in the lungs, which can be mitigated by antibiotic-induced depletion of lung microbiota. These results highlighted the crucial role of lung microbiota in the progression of silica-induced lung fibrosis.
-
A stacked bar chart showing the composition of the lung microbiota in each group at the phylum level is presented in the Figure 5A. At the genus level, differences in microbial composition are depicted in a heatmap (Figure 5B), illustrating the distinct microbiota profile of the silica group. This group exhibited an increased abundance of Acinetobacter, Deinococcus, Micrococcus, Citricoccus, Brachybacterium, and Brevundimonas, among others compared to the other three groups. The LEfSe analysis (Supplementary Table S1) further demonstrated that the phyla Thermi and Actinobacteria were significantly enriched in the SiO2 group compared to the other three groups (Figure 5C). MetagenomeSeq analysis was used to identify distinct ASVs exhibiting relative enrichment in the SiO2 group compared to the PBS and Abx+SiO2 groups (Supplementary Table S2). The intersection of these results revealed 50 shared differential ASVs, which were further classified into 25 different genera, as illustrated in Figure 5D and E. These different ASVs may be related to the lung lesions induced by silica.
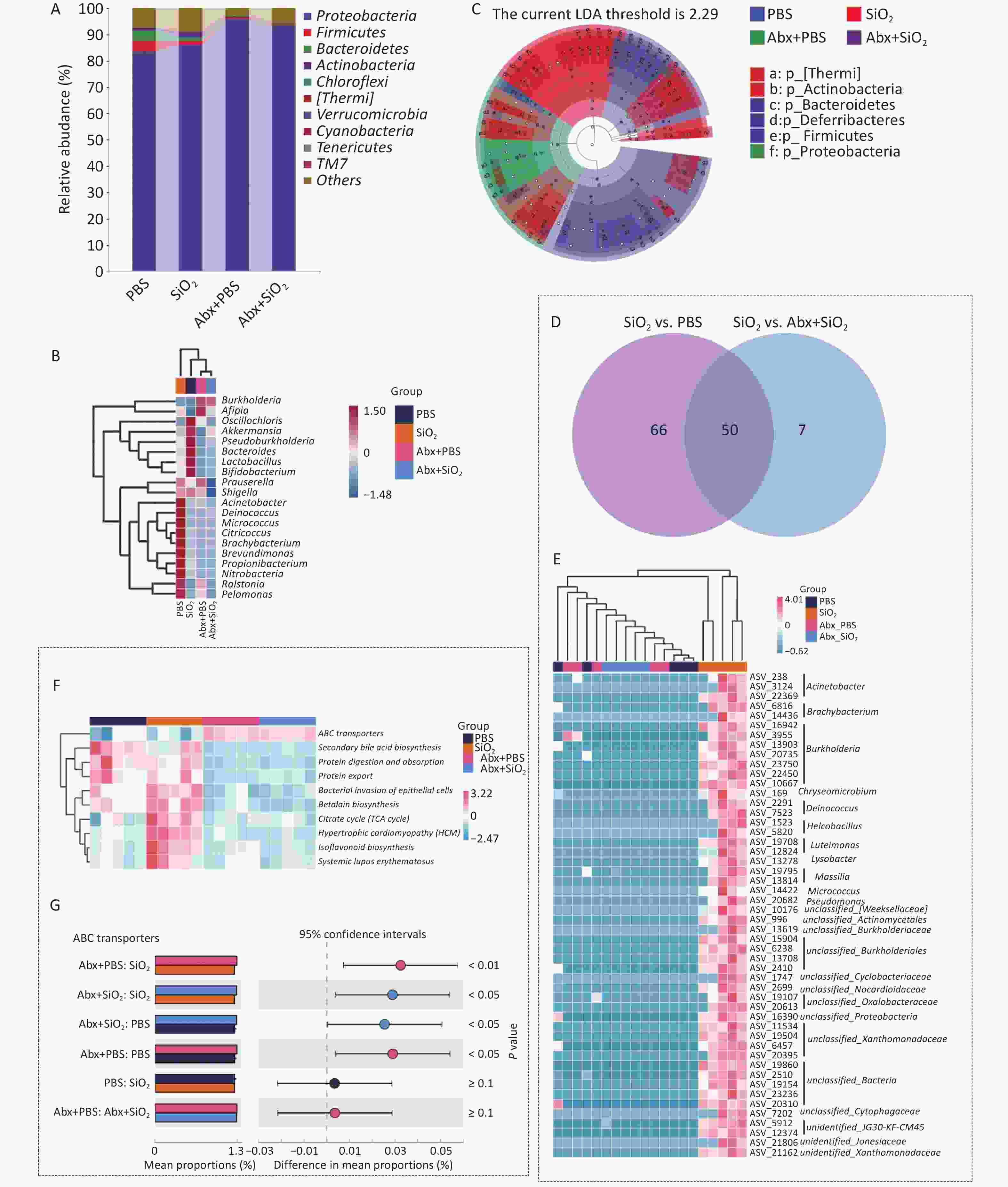
Figure 5. Alterations in the composition of the lung microbiota in the bronchoalveolar lavage fluid (BALF) of mice with Abx pretreatment and/or silica exposure. (A) The top 10 most abundant phyla in the lung microbiota. (B) Heatmap illustrating the top 10 differentially abundant genera. (C) LEfSe analysis identifying distinct taxonomic biomarkers among the four groups. (D) Venn diagram and (E) Heatmap of differential bacterial genera depicting changes in lung microbiota based on pairwise comparisons. (F) Predicted functional pathways of pulmonary commensal bacteria of different groups at Level 3 of the KEGG pathway. (G) Bar plot showing the relative abundance of ABC transporters pathway in different groups.
The results of the PICRUSt2 functional prediction further revealed that at KEGG level 3, the SiO2 group exhibited a higher abundance trend in the citrate cycle, hypertrophic cardiomyopathy, and isoflavonoid biosynthesis compared to both the PBS and Abx+SiO2 groups (Figure 5F). Additionally, the SiO2 group exhibited a lower abundance of ABC transport proteins compared to that of the Abx+SiO2 group (Figure 5F and G).
These findings indicate that silica exposure disrupts lung microbiota, leading to an increased relative abundance of commensal bacteria related to silicosis. However, prior antibiotic treatment, which depletes the lung microbiota before silica exposure, alters the relative abundance of these differential commensal bacteria, mitigating silica-induced pulmonary dysbiosis.
-
Combined with the results shown in Figure 4B–E, we further explored whether this pathological difference, due to changes in the lung microbiota caused by antibiotics, was related to SPs expression in mouse lung tissue. The results revealed a significant increase in the expression levels of SP-A and SP-D in the Abx+PBS group compared with those in the PBS group (Figure 6A and B). However, no significant difference was observed in SPs expression levels between the Abx+SiO2 and SiO2 groups. Moreover, while the number of ABCA3-positive cells did not show a significant change, the Abx+SiO2 group exhibited smaller cell nodules than the SiO2 group (Figure 6C). Accordingly, we concluded that the lung microbiota depletion induced by Abx, although it did not trigger significant changes in the physiological outcome of mouse lung tissues, upregulated the upregulation of the expression of SP-A and SP-D in mouse lung tissues, which act as a kind of host defense proteins to alleviate to a certain extent the subsequent silica stimulation-induced inflammation and fibrosis in the lungs.
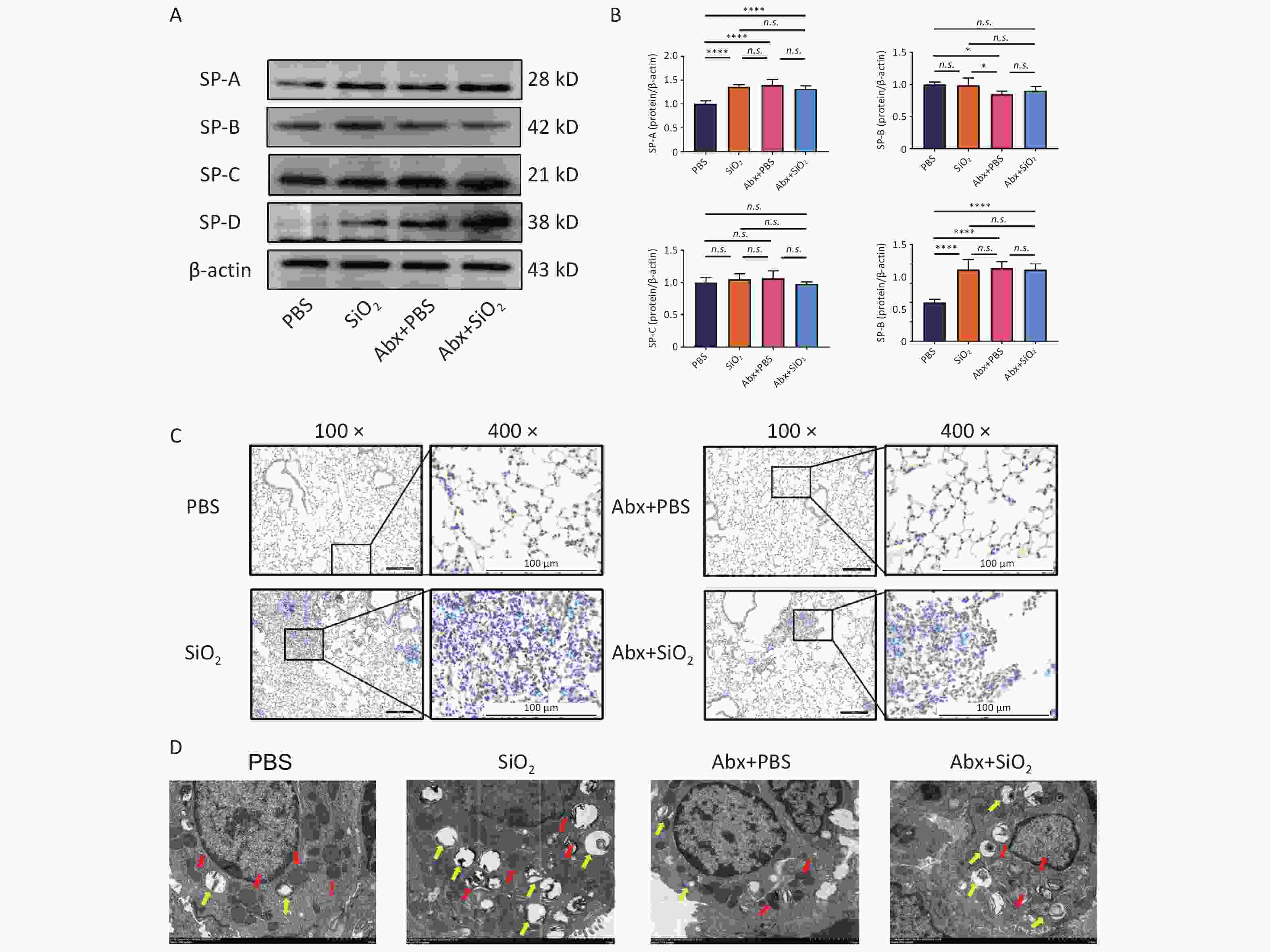
Figure 6. Effects of silica exposure and/or antibiotics administration on surfactant proteins (SPs). (A) Expression levels of SP-A, -B, -C, and -D in the lung tissue of each group, as measured by Western blotting. (B) Corresponding densitometry data for SP-A, -B, -C, and -D protein levels were quantified using ImageJ Software. Bar graphs are presented as the mean ± SD (n = 5). (C) Representative photomicrographs of ABCA3 immunohistochemical staining in the lung tissue of mice in different groups. Scale bars: 100 μm. (D) Morphological changes of the mitochondria and lamellar bodies (LBs) in the ATII cells of mice in each group. Red arrows indicate mitochondria, and yellow arrows indicate LBs. Scale bars: 2 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. indicates no statistically significant difference.
The lung microbiome plays a crucial pathogenic role in bleomycin-induced pulmonary fibrosis and chronic inflammation induces dysbiosis by modifying the oxidative and metabolic milieu of the airways. In this study, the results showed differences between the microbial communities and three histological scores or disease-related proteins in each group at the genus level when performing RDA. Among them, the genera were associated with the three histological scores (inflammation score, IS; fibrosis score, FS; ratio of fibrosis collagen/tissue area, FA) in samples from four groups, with a degree of 98.47% (Supplementary Figure S1A). After 999 permutation tests, 14 genera were found to have significant constraints on the three histological scores. The genera showed 67.44% correlation with the disease-related proteins (Supplementary Figure S1B). Nine genera significantly affected the collagen production and surfactant protein secretion. (Supplementary Table S3).
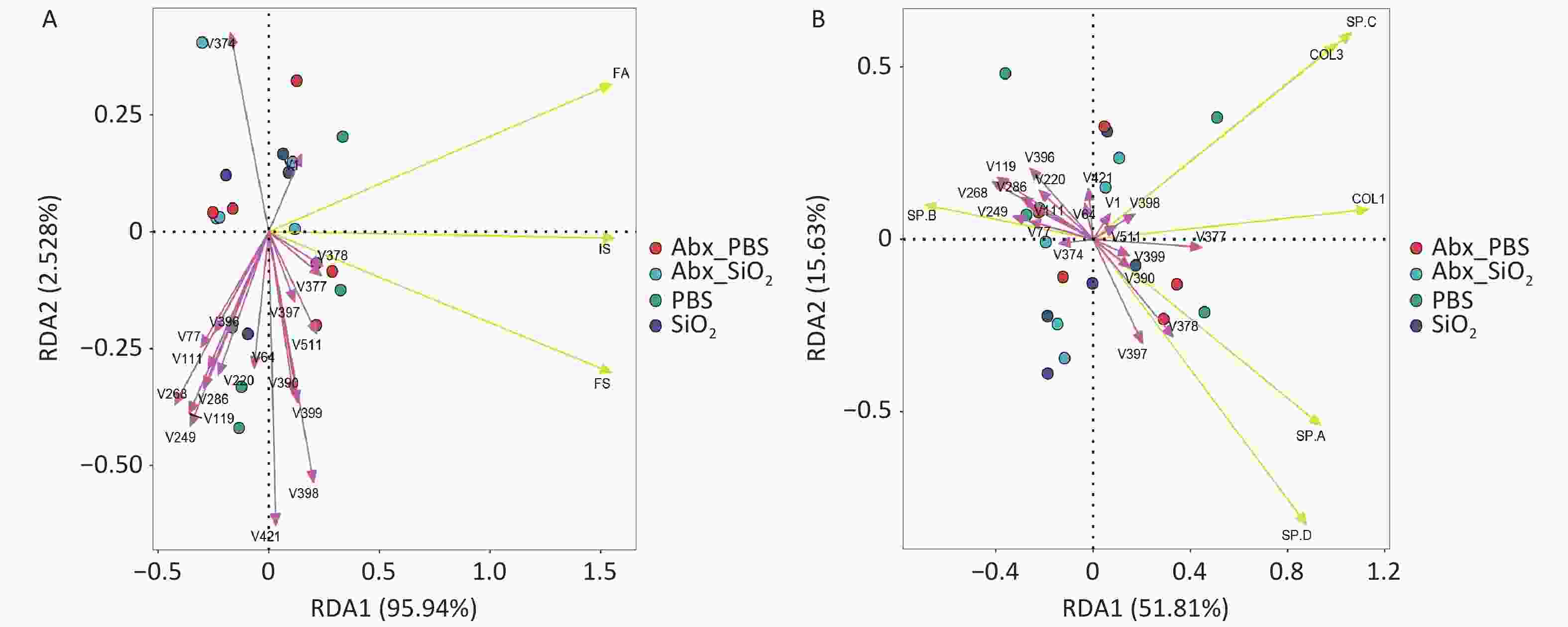
Figure S1. Redundancy analysis (RDA) plots. PCA revealed the correlations between flora and IS (inflammation score), FS (fibrosis score), FA (proportion of fibrosis/ tissue area) (A) or the expression of the disease related proteins COL1 (collagen I), COL3 (collagen III), SP-A, SP-B, SP-C, SP-D, at genus level (B). Each dot indicates one sample; bacteria are indicated by red arrows (The name of the bacterium can be found in Supplementary Table S3), and phenotypic data are indicated by yellow arrows. The length of the arrow line represents the correlation between the bacterial distribution and the phenotypic data, the longer the arrow line, the higher the correlation. The angle between the arrow line and the ranking axis represents the correlation between them, the smaller the angle, the higher the correlation, and vice versa, the lower the correlation.
Furthermore, we used transmission electron microscopy to examine the effects of combined antibiotic pretreatment and silica exposure on alveolar epithelial cells. The results exhibited that, compared to the PBS group, Abx treatment increased the number of mitochondria type II alveolar epithelial cells. However, after silica exposure, the mitochondria appeared enlarged, distorted, and fragmented with reduced cristae. Additionally, silica stimulation led to an increase in lamellar bodies (LBs) volume and accumulation of LBs in ATII cells. Notably, compared with the Abx+SiO2 group, SiO2 group exhibited a significant reduction in LB content, as illustrated in Figure 6D. Combined with the results shown in Figure 4B–4E, these findings further demonstrate that Abx-induced lung microbiota depletion may modulate the response of alveolar epithelial cells to silica-induced injury, mitigating the progression of lung fibrosis through the upregulation of SP-A and SP-D.
-
Silicosis is a lung disease caused by long-term inhalation of silica dust, characterized by diffuse lung inflammation and interstitial fibrosis[25]. Recent studies on the relationship between commensal bacteria and host health have revealed significant differences in the gut microbiota of individuals with respiratory diseases and healthy individuals[26,27]. Additionally, experimental study on silicosis and lung microbiota has demonstrated that the pulmonary microbiota is not only closely associated with pulmonary fibrosis but also plays a crucial role in mitigating the condition. Despite these findings, the interplay between the lung microbiota and SPs secreted by ATII cells during disease progression remains unclear. ATII cells and their secreted SPs are vital for repairing alveolar structures and maintaining immune homeostasis[28,29]. Investigating the relationship between the lung microbiota and SPs in silicosis-related pathology could provide new insights into disease mechanisms and potential therapeutic strategies.
Pulmonary surfactant has long been recognized as intricately involved in the pathogenesis of silicosis[15,30]. Several studies have shown that pulmonary surfactants exerts a protective effect against silica toxicity, and this effect is not confined to specific components of the surfactant, such as phospholipids or SPs[31]. Moreover, SPs, essential components of the pulmonary surfactant system, contribute to lung immunity and host defense against microbial pathogens[32,33]. SP-A and SP-D, in particular, exhibit antimicrobial properties and facilitate microbial clearance by enhancing phagocytosis and opsonization[34]. Our results (Figure 1E and F), suggest that the increased expression SP-A and SP-D may represent as a normal host response to protect against lung injury following silica exposure, consistent with earlier studies in animal models[35].
Lung microbiota has been implicated in the inflammatory response associated with the progression of pulmonary fibrosis. Consistent with our results, a recent study indicated that dysbiosis of the lung microbiota is involved in silica-induced silicosis fibrogenesis process[9]. Therefore, we hypothesized that the pulmonary microbiota contributes to the progression of pulmonary fibrosis by inducing an inflammatory response[36]. Abx mice has emerged as a common experimental model for investigating the effects of lung microbiota depletion or dysbiosis on various physiological processes, including immunity, metabolism, and disease progression[8,37]. Despite their utility, the effect of Abx treatment on SPs in Abx mice has been overlooked in studies focused on the lung microbiota. In our study, the results depicted in Figure 3D and E demonstrate that Abx-treated mice exhibited similar lung structure and function compared to those in the vehicle group. Notably, the expression of SP-A and SP-D in the lung tissues of Abx mice was higher than that of the vehicle-treated mice. Combined with the findings related to ABCA3 mentioned in Figure 3G, these results suggest that, in the absence of additional external stimuli, the depletion of lung commensal bacteria induced by Abx treatment does not affect the alveolar structure. These findings are consistent with the results of a previous study on the airway microbiome of germ-free mice[38]. However, we cannot directly determine whether the upregulation of SP-A and SP-D expression in mouse lung tissues following antibiotic treatment is due to the direct effects of the antibiotics or is mediated through signals associated with changes in the lung microbiome. Nevertheless based on the finding that the transcript levels of lung surfactant protein-related genes in the offspring of germ-free mice are influenced by perinatal microbial exposure[39], we speculate that the changes in SP-A and SP-D expression in lung tissues following microbiome alteration induced by antibiotic treatment may be regulated by commensal bacteria.
To further investigate the effect of the lung microbiota on silica-induced pulmonary inflammation and fibrosis, we examined the changes in lung microbiota composition and diversity in mice with or without Abx treatment, after either silica or PBS administration. In our study, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria were the predominant phyla in C57BL/6 mice, consistent with previous reports[40]. Furthermore, compared with the other three groups, the SiO2 group exhibited a higher relative abundance of Acinetobacter, Deinococcus, Micrococcus, Citricoccus, Brachybacterium, Brevundimonas, Propionibacterium, Nitrobacteria, Ralstonia, and Pelomonas. Among them, Acinetobacter, Micrococcus, and Brevundimonas are opportunistic pathogens can trigger respiratory infections under certain conditions. Ralstonia, a gram-negative bacterium has been found to be enriched in the lung microbiota of various lung diseases[41,42]. Additionally, consistent with the findings of PM2.5-induced changes in lung microbiota in mice reported by Wang et al.[43], the PICRUSt2 analysis suggested that ABC transport abundance was lower in the SiO2 group than that of in the Abx+SiO2 group. A study on lipopolysaccharide-induced acute lung injury indicated that the significant decrease in the ABC transporter pathway observed in the LPS group suggested an adaptation of lung bacteria to inflammatory conditions[44]. Combined with the pathological changes and fibrosis marker proteins in mice across different groups, these findings suggested that the reduction in lung inflammation and fibrosis in silicotic mice following Abx treatment may be associated with alterations in the lung microbiota.
As mentioned above, the expression levels of SPs in lung tissues may be influenced by Abx-induced lung microbiota depletion and silica exposure. To further explore the role of SPs in silicotic mice pre-treated with Abx, we assessed the expression levels of SPs and ABCA3 to investigate the expression and transport of SPs in the lung tissues of mice with or without Abx treatment, following either silica or PBS administration. The results for ABCA3, depicted in Figure 1G and 6C, suggest that the increased number of ABCA3 positive cells under silica stimulation may be a mechanism for maintaining normal alveolar structure and function. Similar to some previous studies on silicotic animals, the expression levels of SP-A and SP-D were significantly increased in the SiO2 group compared with that of the PBS group[45,46]. Notably, differences in SPs expression between the PBS and Abx+PBS groups suggested that Abx treatment enhanced the expression of all four SPs in mouse lung tissue to some extent. Furthermore, considering the minimal difference in SPs expression levels between the SiO2 and Abx+SiO2 groups, this suggests that the upregulation of SP-A and SP-D induced by Abx-mediated lung microbiota depletion may enhance the resistance of mouse lung tissue to silica-induced injury.
Taken together, this study provides insights into the association among the lung microbiota, SPs, and pulmonary fibrosis resulting from silica exposure. The findings reveal that silica-induced lung inflammation and fibrosis are linked to increased expression of SP-A and lung microbiota dysbiosis. Additionally, Abx-induced lung microbiota depletion leads to increased SP-A and SP-D expression, suggesting a potential link between microbial dysbiosis and pulmonary physiology. Abx treatment prior to silica exposure in mice resulted in elevated SP-A and SP-D expression, potentially enhancing resistance to silica-induced lung injury[47]. These findings underscore the pivotal role of the lung microbiota in the progression of silica-induced lung fibrosis and suggest potential therapeutic avenues targeting the relationship between the lung microbiota and SPs. While this study offers promising clinical implications for the managing of silica-induced lung fibrosis, it also highlights several limitations that warrant further research and validation in human populations. A comprehensive understanding of the underlying mechanisms, alongside careful consideration of potential clinical applications, is essential before implementing antibiotic pretreatment or related interventions in the management of silicosis.
全文HTML
Crystalline Silica and Animals
The First Sub Experiment: Murine Model of Silicosis
The Second Subexperiment: A Model of Lung Microbiota Depletion Induced by Antibiotics Treatment
The Third Subexperiment: The Effect of Antibiotic-induced Lung Microbiota on Experimental Silicosis
Collection of Biological Samples
Histological Staining and Evaluation
Assessment of Microbiome Depletion
Real-Time Quantitative PCR (qPCR)
Western Blotting
Routine Transmission Electron Microscopy (TEM)
16S rRNA Sequencing
Statistical Analysis
Pathological Lung Changes and SP Alterations in Mice Exposed to Silica
Silica Exposure Induces the Dysbiosis of Lung Microbiota
Lung Microbiota is Involved in Regulating the Expression of Surfactant Proteins in Mice
Lung Microbiota is Involved in the Pulmonary Inflammation and Fibrosis Induced by Silica
Effects of Silica Exposure and Antibiotics Treatment on Lung Microbiota
Antibiotics-induced Microbiota Depletion Enhances Resistance to Silica-induced Pulmonary Fibrosis in Mice by Up-regulating the Expression of Surfactant Proteins
 24154+Supplementary table 3.xls
24154+Supplementary table 3.xls
|

|
 24154+Supplementary table 2.xls
24154+Supplementary table 2.xls
|

|
 24154+Supplementary table 1.xls
24154+Supplementary table 1.xls
|

|
 24154+Supplementary Materials.pdf
24154+Supplementary Materials.pdf
|

|



 下载:
下载:



 Quick Links
Quick Links